To the editor:
BCR-ABL1 kinase domain (KD) mutations are the most common known cause of treatment failure in chronic myeloid leukemia (CML). Emerging evidence suggests that compound mutations (>1 KD mutation in the same molecule) confer resistance to ponatinib1,2 and combination therapy (GNF-5/nilotinib).3 Several recent studies, including 2 published in Blood, employed nested polymerase chain reaction (PCR) amplification of the BCR-ABL1 KD, followed by cloning and Sanger sequencing4 or next-generation sequencing,5,6 and found a high incidence of compound mutations in imatinib-resistant CML patients with multiple KD mutations. These studies would imply that even a combination approach to therapy would be futile in this setting. Furthermore, they argue strongly against the sequential use of different tyrosine kinase inhibitors in high-risk settings. Surprisingly, however, in most cases reported, the same mutations were found both as compound mutations and as individual mutations in the same patient,4-6 suggesting that the same nucleotide substitution occurred independently multiple times within an individual patient. This complexity is difficult to explain phylogenetically. Based on extensive evidence that PCR frequently mediates recombination between highly similar templates and generates chimeric amplicons containing sequence from >1 different alleles,7-9 we argue that the mutant complexity reported may be inflated due to PCR artifacts.
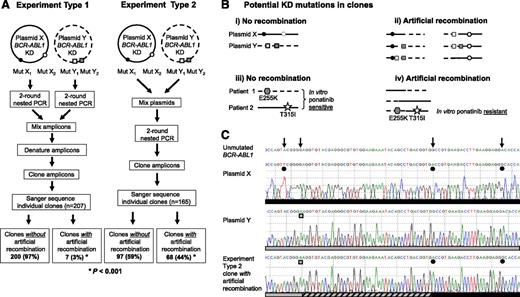
We replicated published procedures4 using mock samples created by mixing mutant BCR-ABL1 plasmids or patient samples, mimicking patients with >1 polyclonal mutant (Figure 1A). When plasmids were PCR amplified singly, and the amplicons of 2 plasmids were mixed, denatured, and cloned (experiment type 1), sequencing of individual clones revealed KD mutations that largely resembled those present in either of the starting plasmids. However, when the plasmids were mixed before PCR amplification (experiment type 2), a large proportion of the resultant clones had KD mutations that originated from both of the starting plasmids (depicted in Figure 1B-C), suggesting that recombination had occurred during PCR amplification. We repeated this using 7 mixtures of 5 different plasmids and found that 20% to 67% of clones showed evidence of artificial recombination resulting in compound mutations that were not present in the starting material, compared with 3% in the control experiments (P < .001).
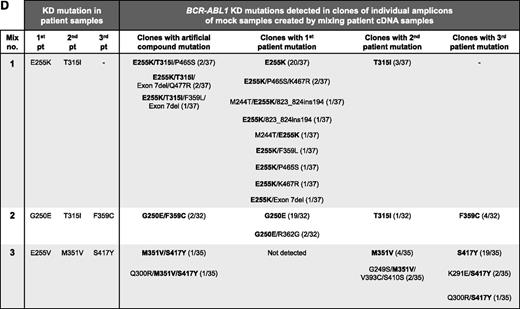
PCR artifacts may mimic BCR-ABL1 compound mutations in CML patients. (A) Outline of the experimental procedure used. Experiment type 1 controls for recombination during cloning into Escherichia coli; mutant BCR-ABL1 plasmids were subjected to PCR amplification individually, and then equal quantities of amplicons of 2 plasmids were mixed, denatured by heating to 95°C for 5 minutes, and cloned. Experiment type 2 mimics amplification of samples from patients with ≥2 mutant BCR-ABL1 clones; equal quantities of 2 plasmids were mixed and subjected to PCR amplification and cloning. Both types of experiments were replicated using 7 mixtures of 5 different plasmids, each containing 4 to 10 KD mutations. Approximately 3000 copies of each plasmid were used as template for each PCR. Unless otherwise specified, first-round PCR (40 cycles) was performed using the Roche Expand Long Template PCR System and the primers 5′-TGACCAACTCGTGTGTGAAACTC-3′ and 5′- TTCGTCTGAGATACTGGATTCCTG-3′, generating ∼1.5 kb amplicons. After cleanup with ExoSAP-IT (Affymetrix), 1 μL of the amplicons was used as template in a second-round PCR (40 cycles) using the primers 5′-GGGCTCTATGGGTTTCTGAATG-3′ and 5′-ATACTGGATTCCTGGAACATTGTTT-3′, generating ∼1.5 kb amplicons containing the BCR-ABL1 KD. Amplified fragments were cloned into pGEM-T Easy (Promega, Madison, WI) and transformed into E coli strain JM109 to minimize E coli–mediated recombination and repair of heterologous DNA. Individual clones (14-37 per mixture) were subjected to Sanger sequencing to reveal the BCR-ABL1 KD sequence within individual amplicons. Clones without artificial recombination are those where the KD mutations resemble those in either of the plasmids in the mixture, and conversely clones with artificial recombination are those with KD mutations originating from both of the plasmids in the original mixture. (B) The KD mutations potentially generated in clones if artificial recombination did not (i, iii) or did (ii, iv) occur. Circles represent compound mutations in plasmid X, and boxes represent compound mutations in plasmid Y; hexagons represent E255K (mix 1, first patient), and stars represent T315I (mix 1, second patient). (C) Sanger sequencing chromatograms showing KD mutations present in a representative plasmid X (black circles) and plasmid Y (gray boxes) and a clone generated using the experiment type 2 procedure. This clone contained 2 of 3 mutations originating from plasmid X as well as 1 mutation originating from plasmid Y. The artificial recombination event occurred within the region marked by the hashed bar. (D) Artificial BCR-ABL1 compound mutations are generated by PCR amplification of mock samples created by mixing equal quantities of cDNA from 8 different CML patients (analogous to experiment type 2; 3 mixtures of 2-3 patient cDNA samples each). Where human tissue was involved, research was conducted with institutional ethics review board approval and in conformance with the Declaration of Helsinki. Mutations present in the original patient samples are shown in bold. Mutations present in >1 clone, but not detected by Sanger sequencing or mass spectrometry in the individual patient samples (“additional mutations”), are shown in regular text; with the exception of exon 7 deletion, these mutations have not been reported in CML patients and likely represent artifacts generated by inaccurate nucleotide incorporation by the DNA polymerase. The frequency of each clone is shown in proportion to the number of clones sequenced per mixture. A total of 13 different clones were detected for mix 1, 5 different clones for mix 2, and 7 different clones for mix 3.
PCR artifacts may mimic BCR-ABL1 compound mutations in CML patients. (A) Outline of the experimental procedure used. Experiment type 1 controls for recombination during cloning into Escherichia coli; mutant BCR-ABL1 plasmids were subjected to PCR amplification individually, and then equal quantities of amplicons of 2 plasmids were mixed, denatured by heating to 95°C for 5 minutes, and cloned. Experiment type 2 mimics amplification of samples from patients with ≥2 mutant BCR-ABL1 clones; equal quantities of 2 plasmids were mixed and subjected to PCR amplification and cloning. Both types of experiments were replicated using 7 mixtures of 5 different plasmids, each containing 4 to 10 KD mutations. Approximately 3000 copies of each plasmid were used as template for each PCR. Unless otherwise specified, first-round PCR (40 cycles) was performed using the Roche Expand Long Template PCR System and the primers 5′-TGACCAACTCGTGTGTGAAACTC-3′ and 5′- TTCGTCTGAGATACTGGATTCCTG-3′, generating ∼1.5 kb amplicons. After cleanup with ExoSAP-IT (Affymetrix), 1 μL of the amplicons was used as template in a second-round PCR (40 cycles) using the primers 5′-GGGCTCTATGGGTTTCTGAATG-3′ and 5′-ATACTGGATTCCTGGAACATTGTTT-3′, generating ∼1.5 kb amplicons containing the BCR-ABL1 KD. Amplified fragments were cloned into pGEM-T Easy (Promega, Madison, WI) and transformed into E coli strain JM109 to minimize E coli–mediated recombination and repair of heterologous DNA. Individual clones (14-37 per mixture) were subjected to Sanger sequencing to reveal the BCR-ABL1 KD sequence within individual amplicons. Clones without artificial recombination are those where the KD mutations resemble those in either of the plasmids in the mixture, and conversely clones with artificial recombination are those with KD mutations originating from both of the plasmids in the original mixture. (B) The KD mutations potentially generated in clones if artificial recombination did not (i, iii) or did (ii, iv) occur. Circles represent compound mutations in plasmid X, and boxes represent compound mutations in plasmid Y; hexagons represent E255K (mix 1, first patient), and stars represent T315I (mix 1, second patient). (C) Sanger sequencing chromatograms showing KD mutations present in a representative plasmid X (black circles) and plasmid Y (gray boxes) and a clone generated using the experiment type 2 procedure. This clone contained 2 of 3 mutations originating from plasmid X as well as 1 mutation originating from plasmid Y. The artificial recombination event occurred within the region marked by the hashed bar. (D) Artificial BCR-ABL1 compound mutations are generated by PCR amplification of mock samples created by mixing equal quantities of cDNA from 8 different CML patients (analogous to experiment type 2; 3 mixtures of 2-3 patient cDNA samples each). Where human tissue was involved, research was conducted with institutional ethics review board approval and in conformance with the Declaration of Helsinki. Mutations present in the original patient samples are shown in bold. Mutations present in >1 clone, but not detected by Sanger sequencing or mass spectrometry in the individual patient samples (“additional mutations”), are shown in regular text; with the exception of exon 7 deletion, these mutations have not been reported in CML patients and likely represent artifacts generated by inaccurate nucleotide incorporation by the DNA polymerase. The frequency of each clone is shown in proportion to the number of clones sequenced per mixture. A total of 13 different clones were detected for mix 1, 5 different clones for mix 2, and 7 different clones for mix 3.
No hot-spot regions were observed for recombination events. Recombination events occurred at similar frequency using several DNA polymerases: 39% with Roche Expand Long (9/23 clones showed artificial recombination), 29% with Roche FastStart (7/24 clones), and 38% with NEB Q5 (13/34 clones). Single-round PCR (35 cycles) using Roche Expand Long significantly reduced recombination events compared with nested PCR using the same enzyme (0% [0/21]; P = .0016).
The procedure was replicated with 3 mock samples created by mixing complementary DNA (cDNA) from 8 patients, each with 1 KD mutation detected by direct Sanger sequencing and sensitive mass spectrometry.10 Artificial compound mutations were detected in clones of all mixtures, including E255K/T315I compound mutation predicted to confer resistance to ponatinib (Figure 1D). Additional mutations, not present in any of the patient samples, were detected in some clones, suggesting that inaccurate nucleotide incorporation by the DNA polymerase also contributes to artifact mutations.
Our study demonstrates that PCR artifacts may mimic BCR-ABL1 compound mutations, leading to inaccurate assessment of mutation status, which could have serious clinical consequences for patients. We urge caution when interpreting results using current procedures and call for new techniques to more reliably detect compound mutations and differentiate them from multiple polyclonal mutations. This will enable r7ational adjustment to the therapeutic approach and more accurate assessment of the impact of various mutations on patient outcome.
Authorship
Acknowledgments: This work was supported by National Health and Medical Research Council of Australia grant 1027531 (S.B., H.S.S., and T.P.H.) and fellowship 1023059 (H.S.S.), a Leukaemia Foundation of Australia/Cure Cancer Australia postdoctoral fellowship (W.T.P.), and a Leukaemia Foundation of Australia and AR Clarkson PhD scholarship (D.T.O.Y.). The Centre for Cancer Biology is an alliance between SA Pathology and the University of South Australia.
Contribution: W.T.P. contributed to experimental design, performed research, analyzed data, and wrote the manuscript; S.R.P. performed research; D.T.O.Y., H.S.S., and S.B. contributed to experimental design and manuscript preparation; and T.P.H. contributed to manuscript preparation.
Conflict-of-interest disclosure: S.B. and T.P.H. receive research funding and honoraria from Novartis Pharmaceuticals, Bristol-Myers Squibb, and Ariad Pharmaceuticals. D.T.O.Y. receives research funding and honoraria from Novartis Pharmaceuticals and Bristol-Myers Squibb. The remaining authors declare no competing financial interests.
Correspondence: Wendy Parker, Department of Genetics and Molecular Pathology, Centre for Cancer Biology, SA Pathology, Frome Rd, Adelaide SA 5000, Australia; e-mail: wendy.parker@health.sa.gov.au.