Key Points
This study deciphers the regulatory role of miR-146a during GVHD in mice.
In humans, the minor genotype of the SNP rs2910164, which reduces expression of miR-146a, was associated with higher GVHD severity.
Abstract
Acute graft-versus-host disease (GVHD) limits the success of allogeneic hematopoietic cell transplantation (allo-HCT); therefore, a better understanding of its biology may improve therapeutic options. We observed miR-146a up-regulation in T cells of mice developing acute GVHD compared with untreated mice. Transplanting miR-146a−/− T cells caused increased GVHD severity, elevated tumor necrosis factor (TNF) serum levels, and reduced survival. TNF receptor-associated factor 6 (TRAF6), a verified target of miR-146a, was up-regulated in miR-146a−/− T cells following alloantigen stimulation. Higher TRAF6 levels translated into increased nuclear factor-κB activity and TNF production in miR-146a−/− T cells. Conversely, the detrimental effect of miR-146a deficiency in T cells was antagonized by TNF blockade, whereas phytochemical induction of miR-146a or its overexpression using a miR-146a mimic reduced GVHD severity. In humans, the minor genotype of the single nucleotide polymorphism rs2910164 in HCT donors, which reduces expression of miR-146a, was associated with severe acute GVHD (grade III/IV). We show that miR-146a functions as a negative regulator of donor T cells in GVHD by targeting TRAF6, leading to reduced TNF transcription. Because miR-146a expression can be exogenously enhanced, our results provide a novel targeted molecular approach to mitigate GVHD.
Introduction
Acute graft-versus-host disease (GVHD) limits the success of allogeneic hematopoietic cell transplantation (allo-HCT).1 To diminish the risk of GVHD, allo-HCT patients receive immunosuppressive treatment that delays immune reconstitution, thereby increasing the incidence of relapse and opportunistic infections. A better understanding of the pathophysiology of GVHD may help to make allo-HCT a safer treatment option, especially for patients that are at high risk for GVHD.
Micro-RNAs (miRNAs) have been shown to be potent regulators of various biological processes. Their modification could tune the allogeneic immune response at multiple levels because they often have several target genes. Together with proteins from the Argonaute family, miRNAs form the RNA-induced silencing complex and interact with partially complementary sequences located primarily in the 3′ untranslated region of their target mRNA, resulting in either inhibition of translation or degradation of the mRNA. Because miRNAs need only to partially pair with the target mRNA to elicit translational repression, a single microRNA can target several genes, whereas a single protein-coding mRNA is often regulated by multiple microRNAs. We recently showed that miR-100 can regulate inflammatory neovascularization during GVHD.2 Because miR-100 expression is normally confined to endothelial cells and not immune cells, other miRNAs that directly down-modulate immune responses such as miR-146a/miR-146b3,4 and miR-124,5 or enhance inflammation such as miR-1556 and miR-142,7 could be potent checkpoints to regulate GVHD.
miR-146a controls innate immune cell and T-cell responses, and its deficiency causes autoimmunity.3 Mechanistically, miR-146a has been demonstrated to directly target 2 adapter proteins in the nuclear factor (NF)-κB activation pathway, tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6) and interleukin (IL)-1 receptor-associated kinase 1, both in innate immune cells3,8 and T cells.9 miR-146a gene expression analysis has demonstrated induction in response to microbial components such as lipopolysaccharide (LPS),8 which trigger GVHD pathology.10 In addition, the survival and maturation of human plasmacytoid dendritic cells that are involved in GVHD11 were shown to be regulated by miR-146a.12 Hence, this miRNA could contribute to the pathogen-mediated inflammation during GVHD initiation, which we have recently shown to be a key event in early GVHD pathogenesis.13 In addition, FoxP3+ regulatory T cells (Tregs) suppress GVHD14 and have been shown to be dependent on miR-146a for the control of Th1 responses and associated autoimmunity.15
Therefore, we investigated the role of miR-146a in donor T cells in an allo-HCT setting and observed that miR-146a negatively regulates GVHD via targeting of TRAF6, leading to reduced TNF transcription in allogeneic T cells. We further analyzed hematopoietic cell donors for a single nucleotide polymorphism (SNP) within the pre-miR-146a sequence and found an association of the donor CC genotype with severe acute GVHD. The minor CC genotype is known to cause decreased miR-146a production. Thus, we provide evidence that miR-146a acts as an important negative regulator in murine and human GVHD and suggest the exogenous increase of miR-146a as a potential novel strategy for therapeutic intervention in this disease.
Materials and methods
Human subjects
For the case-control study, genomic DNA from 286 hematopoietic stem cell donors was isolated to determine their miR-146a rs2910164 genotypes. We collected all human samples after approval by the Ethic Committee of the Albert Ludwigs University Freiburg, Freiburg, Germany (protocol 394/13) and after written informed consent in accordance with the Declaration of Helsinki. GVHD grading was performed on the basis of histopathology as described for human GVHD.16
Mice
C57BL/6 (H-2Kb) and BALB/c (H-2Kd) mice were obtained from the local stock of the animal facility at Freiburg University Medical Center. miR-146a−/− mice on the C57BL/6 background and C.B10-H2b/LilMcdJ mice (referred to as BALB/B) were purchased from Jackson Laboratories. Tnf−/− mice on the C57BL/6 background were kindly provided by the Max-Planck Institute of Immunobiology and Epigenetics, Freiburg, Germany. Mice were housed under specific pathogen-free conditions at the mouse facility of the University Medical Center Freiburg. Only age- and gender-matched mice that were 7 to 12 weeks old were used for transplant experiments. All animal protocols (G-12/34, G-11/36, X-12/02J, and X-13/07J) were approved by the Federal Ministry for Nature, Environment and Consumer Protection of the state of Baden-Württemberg.
BM transplantation model and GVHD histopathology
The majority of mouse bone marrow (BM) transplantation experiments was performed using the C57BL/6 into BALB/c model as previously described.2 Briefly, recipients (BALB/c) were irradiated with 9 Gy, followed by intravenous injection of 5 × 106 allogeneic (C57BL/6) BM cells on day 0. To induce acute GVHD, 2 to 3 × 105 allogeneic (C57BL/6) CD4+/CD8+ T cells isolated by magnetic-activated cell sorting were injected intravenously on day 0. Additional BM transplantation models used are described in the supplemental Methods available on the Blood Web site. Sections of liver, small intestine, and colon collected on day 7 after allo-HCT were stained with hematoxylin and eosin and scored on the basis of a published histopathology scoring system17 by 2 investigators blinded to the experimental groups.
Nucleofection of T cells with miRNA mimics
T cells were purified from wild-type (WT) BALB/c, C57BL/6, or Tnf−/− mice with the Pan T cell Isolation Kit (Miltenyi) before transfection using the Amaxa Mouse T-cell Nucleofector Kit (Lonza) according to the manufacturer's protocol. Briefly, 2 × 106 T cells were resuspended in 100 μL Nucleofector Solution, and 10 pmol of mirVana miRNA mimic Negative Control #1 (Invitrogen) or mirVana miRNA-146a mimic (miRBase ID mmu-miR-146a-5p; Invitrogen) was added to a final concentration of 100 nM. Nucleofector program X-001 was run, and transfected cells were incubated at 37°C/5% CO2 for 4 hours before transplantation.
Microarray analysis
Splenic CD4+/CD8+ T cells (C57BL/6; miR-146a−/− or miR-146a+/+) were stimulated with irradiated (40 Gy) allogeneic BM-derived dendritic cells (BMDCs) (BALB/c) in 6-well plates. After 48 hours, T cells were isolated by negative selection using CD11c microbeads (Miltenyi Biotech) and reached a purity of >85%. RNA was isolated from the purified T cells using the RNeasy Mini Kit (Qiagen), and the sample quality was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies). Microarray was performed using Affymetrix WT Mouse Gene 1.0 ST arrays, as further described in the supplemental Methods.
SNP analysis
Genomic DNA was extracted from peripheral blood samples using the QIAamp DNA Micro Kit and a QIAcube System (Qiagen). The SNP rs2910164 (mature hsa-miR-146a transcript) was analyzed using the real-time polymerase chain reaction (PCR)-based Taqman SNP Genotyping Assay (ASSAY ID C_15946974_10, SNP DB ID: rs2910164; Life Technologies) according to protocols provided by the manufacturer and further described in the supplemental Methods.
Statistical analysis
GraphPad Prism version 5.03 was used for statistical analysis. For normally distributed data, a 2-tailed unpaired Student t test was used. The Mann-Whitney U test was used when the data did not conform to a normal distribution. Differences in animal survival (Kaplan-Meier survival curves) were analyzed by log-rank test.
The Hardy-Weinberg equilibrium for the polymorphism rs2910164 was analyzed using a χ2 test. Frequencies of genotypes were compared using Fisher's exact test. Data are presented as mean ± standard error of the mean. Differences were considered significant when P < .05.
Further materials and methods are described in the supplemental Methods.
Results
miR-146a deficiency in T cells enhances GVHD
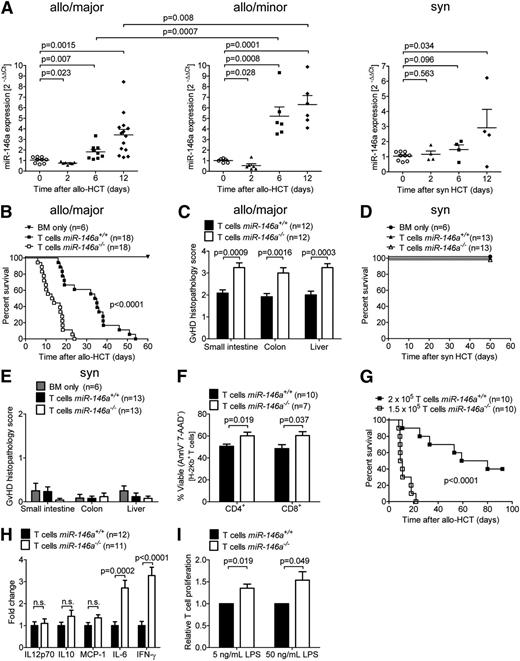
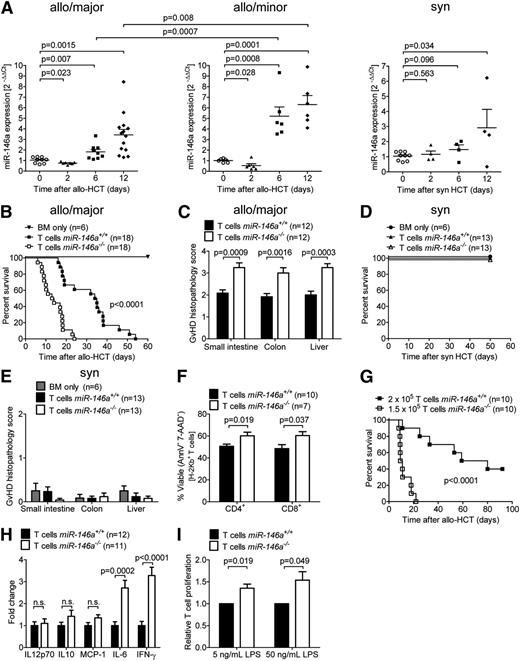
To determine whether miR-146a is differentially regulated at the transcriptional level during GVHD progression in mice, we isolated T cells from spleens and inguinal lymph nodes (LNs) at different time points before (day 0) or after major mismatch allo-HCT and determined miR-146a expression by quantitative reverse transcriptase-PCR (qRT-PCR). miR-146a was significantly up-regulated in the T cells of mice developing acute GVHD (days 6 and 12) compared with untreated mice, whereas it was down-regulated shortly after allo-HCT (day 2), when T cells generally get primed (Figure 1A, left). A similar kinetics for miR-146a regulation was observed after allo-HCT in a minor mismatch model (Figure 1A, center). Because miR-146a functions as a negative regulator of inflammation, its up-regulation (days 6 and 12) in T cells, as the primary effector cells in GVHD, could be interpreted as a rescue mechanism to counteract inflammation. This is consistent with the fact that miR-146a expression was significantly lower on days 6 and 12 after major mismatch allo-HCT than after minor mismatch allo-HCT, a setting with less severe acute GVHD. The down-regulation of miR-146a early (day 2) after allo-HCT did not occur after syngeneic transplantation (Figure 1A, right), indicating that miR-146a is inadequately regulated early after allo-HCT in mice developing GVHD.
miR-146a is regulated in the T-cell compartment after HCT and its deficiency in T cells enhances GVHD. (A) T cells were isolated from the spleens and inguinal LNs of untreated mice (day 0) or on days 2, 6, and 12 after (left) major MHC mismatch allo-HCT (C57BL/6→BALB/c), (center) minor histocompatibility antigen mismatch allo-HCT (BALB/B→C57BL/6), or (right) syngeneic HCT (BALB/c→BALB/c) by magnetic-activated cell sorting enrichment for CD4 and CD8. miR-146a expression was analyzed by qRT-PCR. Each data point represents an individual animal. (B-C) Allo-HCT was performed as described for the C57BL/6 into BALB/c combination. (B) Survival of BALB/c recipient mice transplanted with WT BM cells plus T cells from miR-146a−/− or miR-146a+/+ mice as indicated is shown. (C) The small intestine, colon, and liver were isolated from mice receiving T cells from miR-146a−/− or miR-146a+/+ mice on day 7 after allo-HCT and scored for GVHD severity. (D-E) Syngeneic HCT was performed as described for the C57BL/6 into C57BL/6 model. (D) Survival of C57BL/6 recipient mice transplanted with WT BM cells plus T cells from syngeneic miR-146a−/− or miR-146a+/+ mice as indicated is shown. (E) Small intestines, colons, and livers were isolated on day 50 after syngeneic HCT and scored for GVHD severity. (F-H) Allo-HCT was performed as described for the C57BL/6 into BALB/c combination. (F) T cells were isolated from the spleens of allo-HCT hosts receiving T cells from miR-146a−/− or miR-146a+/+ mice on day 7 after allo-HCT and analyzed for apoptosis/necrosis via Annexin-V/7-AAD staining. (G) Recipient mice receiving 2 × 105miR-146a+/+ T cells or 25% less miR-146a−/− T cells as indicated were monitored for survival. (H) The indicated cytokines were measured in the serum of recipient mice receiving T cells from miR-146a−/− or miR-146a+/+ mice on day 7 after allo-HCT by cytometric bead array. (I) Proliferation of carboxyfluorescein diacetate succinimidyl ester-labeled T cells (C57BL/6) in response to coculture with LPS-stimulated BMDCs (BALB/c) for 96 hours at a 1:1 ratio is shown for T cells derived from miR-146a−/− compared with miR-146a+/+ mice. Data were normalized to the miR-146a+/+ control group. All data were pooled from 2 to 3 independent experiments.
miR-146a is regulated in the T-cell compartment after HCT and its deficiency in T cells enhances GVHD. (A) T cells were isolated from the spleens and inguinal LNs of untreated mice (day 0) or on days 2, 6, and 12 after (left) major MHC mismatch allo-HCT (C57BL/6→BALB/c), (center) minor histocompatibility antigen mismatch allo-HCT (BALB/B→C57BL/6), or (right) syngeneic HCT (BALB/c→BALB/c) by magnetic-activated cell sorting enrichment for CD4 and CD8. miR-146a expression was analyzed by qRT-PCR. Each data point represents an individual animal. (B-C) Allo-HCT was performed as described for the C57BL/6 into BALB/c combination. (B) Survival of BALB/c recipient mice transplanted with WT BM cells plus T cells from miR-146a−/− or miR-146a+/+ mice as indicated is shown. (C) The small intestine, colon, and liver were isolated from mice receiving T cells from miR-146a−/− or miR-146a+/+ mice on day 7 after allo-HCT and scored for GVHD severity. (D-E) Syngeneic HCT was performed as described for the C57BL/6 into C57BL/6 model. (D) Survival of C57BL/6 recipient mice transplanted with WT BM cells plus T cells from syngeneic miR-146a−/− or miR-146a+/+ mice as indicated is shown. (E) Small intestines, colons, and livers were isolated on day 50 after syngeneic HCT and scored for GVHD severity. (F-H) Allo-HCT was performed as described for the C57BL/6 into BALB/c combination. (F) T cells were isolated from the spleens of allo-HCT hosts receiving T cells from miR-146a−/− or miR-146a+/+ mice on day 7 after allo-HCT and analyzed for apoptosis/necrosis via Annexin-V/7-AAD staining. (G) Recipient mice receiving 2 × 105miR-146a+/+ T cells or 25% less miR-146a−/− T cells as indicated were monitored for survival. (H) The indicated cytokines were measured in the serum of recipient mice receiving T cells from miR-146a−/− or miR-146a+/+ mice on day 7 after allo-HCT by cytometric bead array. (I) Proliferation of carboxyfluorescein diacetate succinimidyl ester-labeled T cells (C57BL/6) in response to coculture with LPS-stimulated BMDCs (BALB/c) for 96 hours at a 1:1 ratio is shown for T cells derived from miR-146a−/− compared with miR-146a+/+ mice. Data were normalized to the miR-146a+/+ control group. All data were pooled from 2 to 3 independent experiments.
To clarify this observation with a functional study, we then used gene-targeted mice lacking miR-146a. T cells isolated from miR-146a+/+ or miR-146a−/− mice (H-2Kb) were transplanted into irradiated BALB/c recipient mice (H-2Kd) along with WT BM (H-2Kb). miR-146a−/− T cells caused more aggressive GVHD with significantly enhanced mortality after allo-HCT compared with miR-146a+/+ T cells (Figure 1B; P < .0001). Histological analysis of the small intestine, colon, and liver revealed increased GVHD severity in mice receiving miR-146a−/− T cells compared with miR-146a+/+ T cells (Figure 1C). Conversely, transplantation of syngeneic miR-146a−/− T cells did not cause higher mortality or organ damage in small intestines, large intestines, or liver compared with miR-146a+/+ T cells (Figure 1D-E). As we observed that viability of miR-146a−/− T cells was higher compared with miR-146a+/+ T cells (Figure 1F), we next used 25% fewer miR-146a−/− T cells corresponding to 25% higher viability of miR-146a−/− T cells to counterbalance the lower viability of miR-146a+/+ T cells. Despite the lower number of T cells transferred, the course for GVHD was still more aggressive in the miR-146a−/− T cell group compared with the WT T cell group (Figure 1G).
Serum levels of IL-6 and interferon (IFN)-γ were increased in mice receiving allogeneic miR-146a−/− T cells compared with allogeneic miR-146a+/+ T cells (Figure 1H). Moreover, proliferation of T cells in response to LPS-stimulated allogeneic BMDCs in vitro was higher in T cells derived from miR-146a−/− compared with those from miR-146a+/+ mice (Figure 1I). These data support the concept that miR-146a has a protective role in GVHD and that its absence in the T-cell compartment enhances alloantigen-driven T-cell proliferation and viability, in turn increasing the severity of GVHD.
Exogenous increase of miR-146a expression protects against GVHD
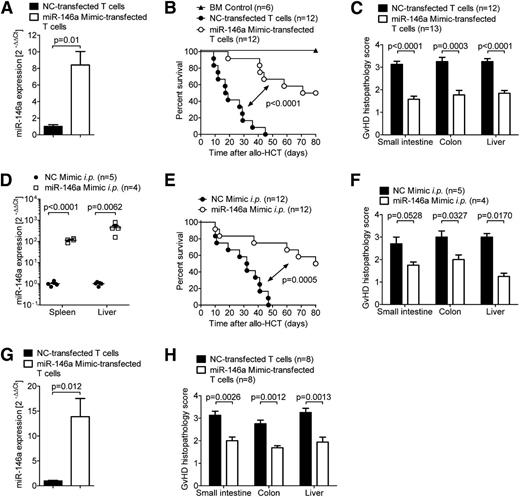
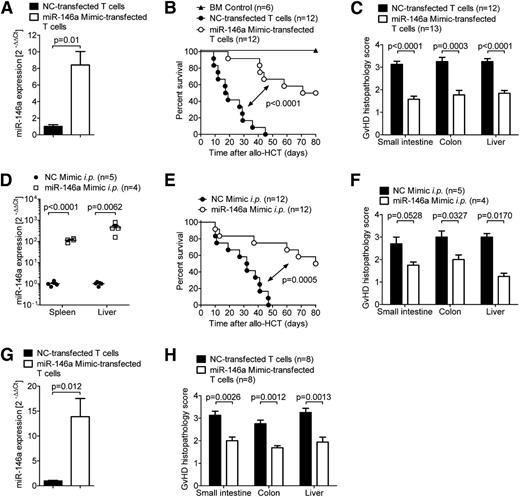
Having shown that miR-146a modulates GVHD, we next investigated whether increased levels of miR-146a could ameliorate GVHD. Therefore, we transfected WT C57BL/6 T cells with a specific miR-146a mimic or a negative control (NC) mimic before transferring them into allogeneic irradiated hosts. Using qRT-PCR, we confirmed that transfection with the miR-146a mimic resulted in more than an eightfold increase of miR-146a levels in cells transfected with the mimic compared with NC (Figure 2A). When the transfected T cells were transferred into irradiated allogeneic recipient mice along with WT BM, we found that mice receiving the miR-146a mimic-transfected T cells exhibited decreased mortality (Figure 2B) and developed less severe GVHD, as assessed by histopathological scoring (Figure 2C). Viability of the miR-146a mimic-transfected T cells was not decreased compared with NC mimic-transfected cells (supplemental Figure 1A). These findings support the concept of a regulatory role for miR-146a in donor T cells.
Exogenously increasing miR-146a levels ameliorates GVHD. (A-C) WT C57BL/6 T cells were transfected with a miR-146a mimic or an NC mimic before transplantation into allogeneic (BALB/c) recipient mice (5 × 105 T cells per mouse). (A) Transfection efficiency was confirmed by qRT-PCR for miR-146a. (B) Survival and (C) GVHD histopathology scores of recipients are depicted. Data were pooled from 2 independent experiments. (D-F) Allo-HCT was performed as described on BALB/c mice using C57BL/6 donors. (D) Mice were treated with the miR-146a mimic or NC mimic in in vivo-jetPEI intraperitoneally on days 2, 5, and 7. miR-146a expression in the spleens and livers of recipient mice was analyzed by qRT-PCR on day 7 after allo-HCT. (E) Survival curves and (F) GVHD histopathology scores are shown. (G-H) WT BALB/c T cells were transfected with a miR-146a mimic or a NC mimic before transplantation into allogeneic (C57BL/6) recipient mice (5 × 105 T cells per mouse). (G) Transfection efficiency was confirmed by qRT-PCR for miR-146a. (H) The small intestine, colon, and liver were isolated on day 7 after allo-HCT and scored for GVHD histopathology.
Exogenously increasing miR-146a levels ameliorates GVHD. (A-C) WT C57BL/6 T cells were transfected with a miR-146a mimic or an NC mimic before transplantation into allogeneic (BALB/c) recipient mice (5 × 105 T cells per mouse). (A) Transfection efficiency was confirmed by qRT-PCR for miR-146a. (B) Survival and (C) GVHD histopathology scores of recipients are depicted. Data were pooled from 2 independent experiments. (D-F) Allo-HCT was performed as described on BALB/c mice using C57BL/6 donors. (D) Mice were treated with the miR-146a mimic or NC mimic in in vivo-jetPEI intraperitoneally on days 2, 5, and 7. miR-146a expression in the spleens and livers of recipient mice was analyzed by qRT-PCR on day 7 after allo-HCT. (E) Survival curves and (F) GVHD histopathology scores are shown. (G-H) WT BALB/c T cells were transfected with a miR-146a mimic or a NC mimic before transplantation into allogeneic (C57BL/6) recipient mice (5 × 105 T cells per mouse). (G) Transfection efficiency was confirmed by qRT-PCR for miR-146a. (H) The small intestine, colon, and liver were isolated on day 7 after allo-HCT and scored for GVHD histopathology.
We next asked whether inducing miR-146a expression in a more therapeutic setting could prevent or delay the development of GVHD after allo-HCT. Therefore, we treated recipient mice intraperitoneally with miR-146a mimic or NC mimic on days 2, 5, and 7 after transplantation, which led to highly increased miR-146a levels in the spleen and liver (Figure 2D). Likewise, treatment with the miR-146a mimic prolonged the survival of recipient mice (Figure 2E) and decreased their histopathological GVHD score (Figure 2F). To analyze the effects of mimic transfection in a second GVHD model, we next used the BALB/c into C57BL/6 model and observed that mice receiving miR-146a mimic-transfected BALB/c T cells developed less severe GVHD, as assessed by histopathological scoring (Figure 2G-H).
To study whether the effects of miR-146a on GVHD could be used therapeutically, we treated the recipient mice with 3,3′-Diindolylmethane (DIM), a phytochemical known to induce miR-146a.18,19 We found that intraperitoneal injection or oral administration of DIM as previously reported20,21 improved survival and decreased GVHD severity in allo-HCT mice (supplemental Figure 1B-D) and augmented miR-146a expression (supplemental Figure 1E-F). Although these results demonstrate the potential of phytochemical therapy to promote tolerogenic miR-146a effects, the precise mechanisms of immune modulation involved are less clear. MiR-146a induction might not be the sole mechanism as to how DIM interferes with GVHD, and it is possible that miR-146a is induced in other cell populations such as donor myeloid cells because miR-146 expression in CD14+ cord blood cells has also been reported.22
miR-146a deficiency leads to increased TNF production in T cells
Next, we aimed to decipher the mechanism as to how miR-146a deficiency renders donor T cells more aggressive in terms of their alloreactivity. To identify functional differences between miR-146a−/− T cells and miR-146a+/+ T cells, we compared the expression of transcription factors that determine T-helper (Th)-1, Th-2, and Th-17 lineage commitment, as well as the Treg numbers and intracellular cytokine profile of T cells isolated from allo-HCT recipient mice. We did not find any differences accounting for the more aggressive behavior of miR-146a−/− T cells, except for a moderate increase in CD44+CD62Llow T cells (supplemental Figure 1A-I). Importantly, the phenotype and the migratory activity of T cells from naïve miR-146a+/+ and miR-146a−/− mice were comparable (supplemental Figure 3A-F). Moreover, immune reconstitution after syngeneic HCT with respect to different lymphoid and myeloid subsets was comparable independent of the transfer of syngeneic miR-146a+/+ and miR-146a−/− T cells (supplemental Figure 4A-H).
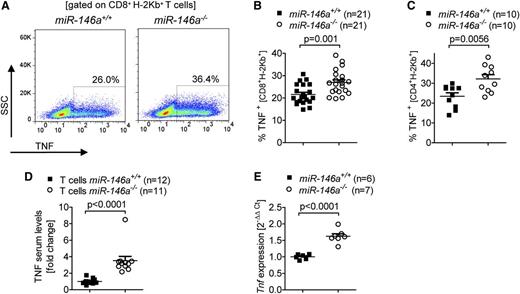
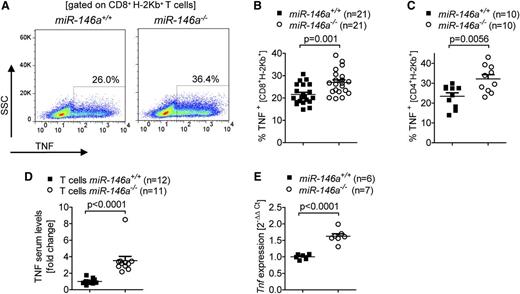
Because previous studies have shown that TNF derived from donor T cells is a critical factor for GVHD induction,23,24 we then asked if TNF production might be altered in miR-146a–deficient T cells. In fact, we detected an increased fraction of TNF-positive miR-146a−/− CD8 and CD4 T cells compared with miR-146a+/+ T cells after the cells were transferred into allogeneic hosts (Figure 3A-C). The higher amount of intracellular TNF in the donor type T cells translated into elevated TNF serum levels when the allo-HCT recipients received miR-146a−/− T cells compared with miR-146a+/+ T cells (Figure 3D). Furthermore, in vitro stimulation of T cells with allogeneic BMDCs caused higher Tnf transcription in miR-146a−/− T cells compared with miR-146a+/+ T cells as determined by qRT-PCR (Figure 3E). These findings demonstrate that, among proinflammatory T cell-derived cytokines, only TNF showed a significant increase in alloantigen exposed miR-146a−/− T cells compared with miR-146a+/+ T cells, which could explain the observed differences in GVHD severity.
TNF levels are increased in miR-146a–deficient T cells. (A-C) The amount of intracellular TNF in miR-146a−/− or miR-146a+/+ CD8+ T cells isolated from allo-HCT recipients on (A-B) day 7 or (C) day 3 is shown. (A) Representative flow cytometry plot gated on CD8+H-2Kb+ T cells. (B) Pooled data from 3 independent transplantation experiments. (C) Pooled data from 2 independent transplantation experiments. (D) TNF levels were measured in the serum of allo-HCT recipient mice receiving T cells from miR-146a−/− or miR-146a+/+ mice on day 7 after transplantation. Data were pooled from 2 independent experiments. (E) CD4+/CD8+ T cells from miR-146a−/− or miR-146a+/+ mice were isolated after 48 hours of stimulation with allogeneic BMDCs (BALB/c) in vitro and reached a purity of >85%. RNA was isolated from purified T cells and Tnf expression was analyzed by qRT-PCR.
TNF levels are increased in miR-146a–deficient T cells. (A-C) The amount of intracellular TNF in miR-146a−/− or miR-146a+/+ CD8+ T cells isolated from allo-HCT recipients on (A-B) day 7 or (C) day 3 is shown. (A) Representative flow cytometry plot gated on CD8+H-2Kb+ T cells. (B) Pooled data from 3 independent transplantation experiments. (C) Pooled data from 2 independent transplantation experiments. (D) TNF levels were measured in the serum of allo-HCT recipient mice receiving T cells from miR-146a−/− or miR-146a+/+ mice on day 7 after transplantation. Data were pooled from 2 independent experiments. (E) CD4+/CD8+ T cells from miR-146a−/− or miR-146a+/+ mice were isolated after 48 hours of stimulation with allogeneic BMDCs (BALB/c) in vitro and reached a purity of >85%. RNA was isolated from purified T cells and Tnf expression was analyzed by qRT-PCR.
Effect of miR-146a deficiency in T cells can be antagonized by TNF blockade
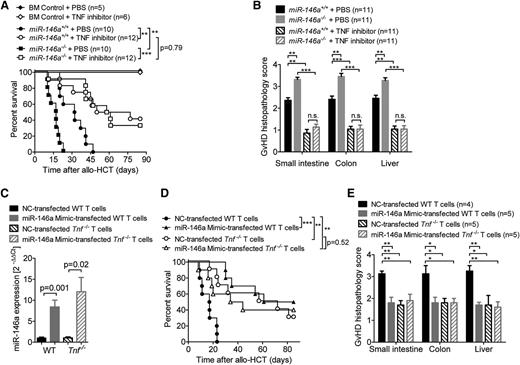
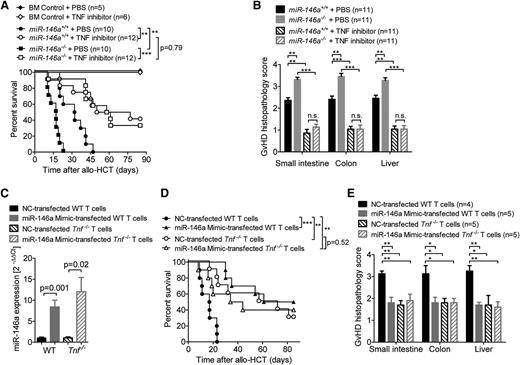
To test whether the observed difference between miR-146a−/− and miR-146a+/+ T cells in their ability to induce GVHD was dependent on TNF production, we used etanercept to block TNF in vivo. When allo-HCT recipients were treated with etanercept, there was no difference in survival between recipients that received miR-146a−/− T cells vs those receiving miR-146a+/+ T cells (Figure 4A). Consistent with this result, there was no difference in histopathological GVHD severity between the 2 recipient groups when etanercept was administered (Figure 4B).
TNF blockade abolishes the detrimental effect of miR-146a deficiency. (A-B) Allo-HCT was performed as described for the C57BL/6 into BALB/c combination. Recipient mice were treated intraperitoneally with the TNF inhibitor etanercept or vehicle (phosphate-buffered saline) 3 times a week for 4 weeks, starting on day 1. The (A) survival and (B) GVHD histopathology scores of BALB/c recipients are shown. Data were pooled from 2 independent experiments. (C-E) WT or Tnf−/− T cells were transfected with a miR-146a mimic or a NC mimic, respectively, before transplantation into allogeneic (BALB/c) hosts (2-5 × 105 T cells per mouse). (C) Transfection efficiency was confirmed by qRT-PCR for miR-146a. The (D) survival (n = 10 per group) and (E) GVHD histopathology scores of recipients are shown. ***P < .0001; **P < .01; *P < .05.
TNF blockade abolishes the detrimental effect of miR-146a deficiency. (A-B) Allo-HCT was performed as described for the C57BL/6 into BALB/c combination. Recipient mice were treated intraperitoneally with the TNF inhibitor etanercept or vehicle (phosphate-buffered saline) 3 times a week for 4 weeks, starting on day 1. The (A) survival and (B) GVHD histopathology scores of BALB/c recipients are shown. Data were pooled from 2 independent experiments. (C-E) WT or Tnf−/− T cells were transfected with a miR-146a mimic or a NC mimic, respectively, before transplantation into allogeneic (BALB/c) hosts (2-5 × 105 T cells per mouse). (C) Transfection efficiency was confirmed by qRT-PCR for miR-146a. The (D) survival (n = 10 per group) and (E) GVHD histopathology scores of recipients are shown. ***P < .0001; **P < .01; *P < .05.
In contrast to TNF inhibition, blockade of IL-6 and IFN-γ did not reverse the more aggressive course of GVHD observed when the donor T cells were derived from miR-146a−/− mice (supplemental Figure 5A-B).
Because etanercept treatment leads to a ubiquitous TNF blockade, we next studied the effects of miR-146a overexpression in T cells that were specifically deficient for Tnf. Therefore, we transfected Tnf+/+ or Tnf−/− T cells with miR-146a mimic before allo-HCT and achieved an 8- to 10-fold increase in miR-146a compared with NC-transfected T cells (Figure 4C). Although Tnf−/− T cells induced less severe GVHD compared with WT T cells, no increase in protection from GVHD was achieved when they were transfected with the miR-146a mimic as measured by survival studies and histopathological GVHD severity scoring (Figure 4D-E). These data indicate that miR-146a overexpression and TNF blockade provide protection via the same mechanism as no synergy could be detected. This provides evidence for a model in which miR-146a negatively regulates the TNF production of alloreactive T cells, leading to reduced GVHD severity.
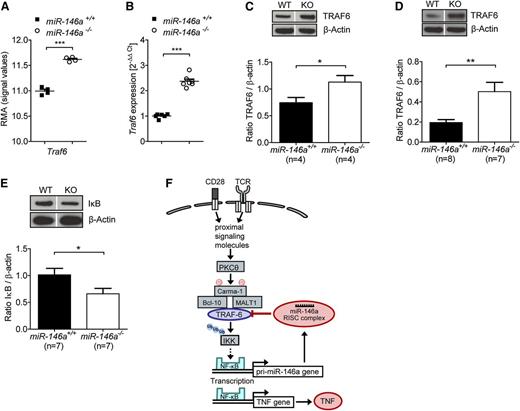
Expression of the miR-146a target TRAF6 is higher in miR-146a–deficient compared with WT T cells on allogeneic stimulation, indicating loss of suppression
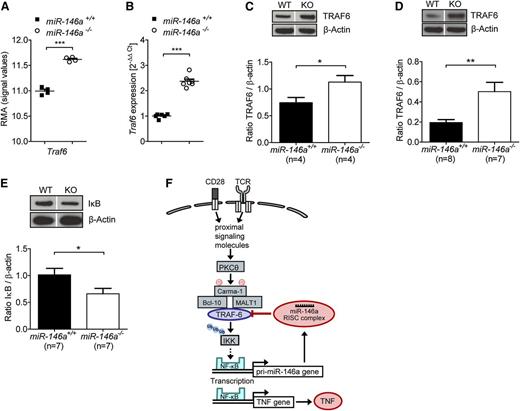
To mechanistically clarify how miR-146a deficiency in donor T cells induces increased TNF levels that exacerbate GVHD, we sought to identify genes that are differentially regulated in miR-146a−/− T cells compared with miR-146a+/+ T cells. Therefore, we used a microarray-based unbiased gene expression approach. RNA was isolated from miR-146a−/− T cells or miR-146a+/+ T cells exposed to allogeneic BMDC in vitro. We checked the most significantly differentially regulated genes against the TargetScan public database (http://www.targetscan.org/) to screen for potential and known targets of mir-146a and to detect the direct effects of miR-146a deficiency (supplemental Figure 6). We identified Traf6 as being both a direct miR-146a target and as being expressed significantly higher in miR-146a−/− T cells, indicating loss of its negative regulation (Figure 5A). This result was intriguing, because TRAF6 is known to be involved in T-cell receptor signaling25 and is one of the best validated targets of miR-146a.8,9
miR-146a deficiency leads to increased TRAF6 expression in T cells stimulated by alloantigen. (A-C) CD4+/CD8+ T cells (C57BL/6) were isolated 48 hours after stimulation with allogeneic BMDC (BALB/c) in vitro. (A) RNA was isolated from purified T cells, and the expression of Traf6 was analyzed by microarray. (B) RNA was isolated from purified T cells and reverse transcribed into cDNA, followed by qRT-PCR. Traf6 expression of T cells isolated from miR-146a−/− or miR-146a+/+ mice is shown. (C) Purified T cells were lysed and analyzed for TRAF6 protein levels by western blot (n = 4 per group). (D) T cells were isolated from the spleens and LNs of allo-HCT recipients on day 9, lysed, and analyzed for TRAF6 expression. Transferred and reisolated T cells were from miR-146a−/− (n = 7) or miR-146a+/+ mice (n = 8) as indicated. (E) The amount of IκB is shown for T cells treated as described in C (n = 7 per group). (F) A proposed simplified mechanism for how the TRAF6/NF-κB/TNF axis is regulated by miR-146a in alloreactive T cells. Dividing lines indicate that lanes were run on the same gel but were noncontiguous. ***P < .0001; **P < .01; *P < .05.
miR-146a deficiency leads to increased TRAF6 expression in T cells stimulated by alloantigen. (A-C) CD4+/CD8+ T cells (C57BL/6) were isolated 48 hours after stimulation with allogeneic BMDC (BALB/c) in vitro. (A) RNA was isolated from purified T cells, and the expression of Traf6 was analyzed by microarray. (B) RNA was isolated from purified T cells and reverse transcribed into cDNA, followed by qRT-PCR. Traf6 expression of T cells isolated from miR-146a−/− or miR-146a+/+ mice is shown. (C) Purified T cells were lysed and analyzed for TRAF6 protein levels by western blot (n = 4 per group). (D) T cells were isolated from the spleens and LNs of allo-HCT recipients on day 9, lysed, and analyzed for TRAF6 expression. Transferred and reisolated T cells were from miR-146a−/− (n = 7) or miR-146a+/+ mice (n = 8) as indicated. (E) The amount of IκB is shown for T cells treated as described in C (n = 7 per group). (F) A proposed simplified mechanism for how the TRAF6/NF-κB/TNF axis is regulated by miR-146a in alloreactive T cells. Dividing lines indicate that lanes were run on the same gel but were noncontiguous. ***P < .0001; **P < .01; *P < .05.
Traf6 changes at the transcriptional level were confirmed by qPCR (Figure 5B) and translated into higher TRAF6 protein levels in T cells exposed to allogeneic dendritic cells in vitro (Figure 5C). Moreover, donor miR-146a−/− T cells isolated from allo-HCT recipient mice on day 9 showed higher TRAF6 protein levels compared with miR-146a+/+ T cells (Figure 5D), thus confirming the results in vivo. Importantly, significantly increased expression of Tnf in miR-146a−/− T cells was also seen in the microarray data (www.ebi.ac.uk/arrayexpress, E-MTAB-2231), confirming our previous results.
TRAF6, an integral component of the T-cell receptor signaling pathway leading to the NF-κB–based transcription of Tnf, mediates the activation of IκB kinase (IKK), which in turn promotes IκB phosphorylation and degradation.25 Because the amount of total IκB is inversely correlated with NF-κB pathway activity, we next studied IκB levels in T cells isolated after in vitro exposure to allogeneic BMDCs. As anticipated, IκB levels were lower in miR-146a−/− T cells compared with miR-146a+/+ T cells (Figure 5E), indicating elevated NF-κB pathway activity after alloantigen stimulation in miR-146a−/− T cells. These results indicate that TRAF6 expression is increased in T cells lacking miR-146a due to a loss of negative regulation translating into higher NF-κB pathway activity.
Based on these findings, we propose that miR-146a down-regulates proinflammatory TNF production in alloreactive T cells by decreasing NF-κB signaling via negative regulation of TRAF6 (Figure 5F).
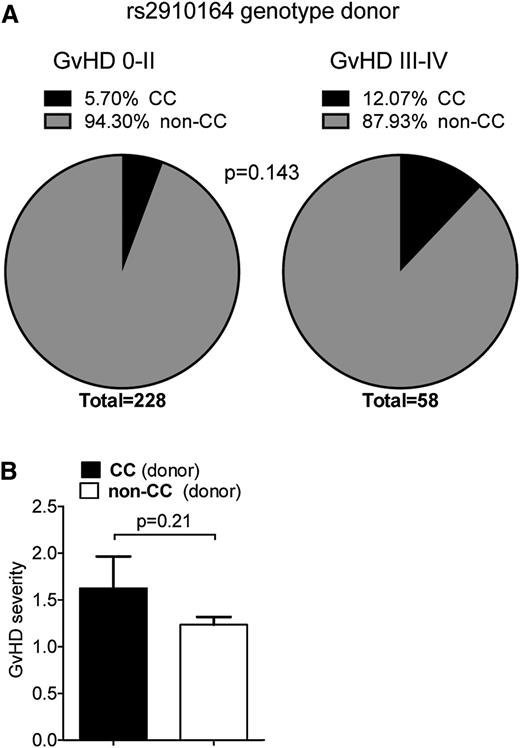
miR-146a SNP rs2910164 genotype is associated with GVHD severity in allo-HCT patients
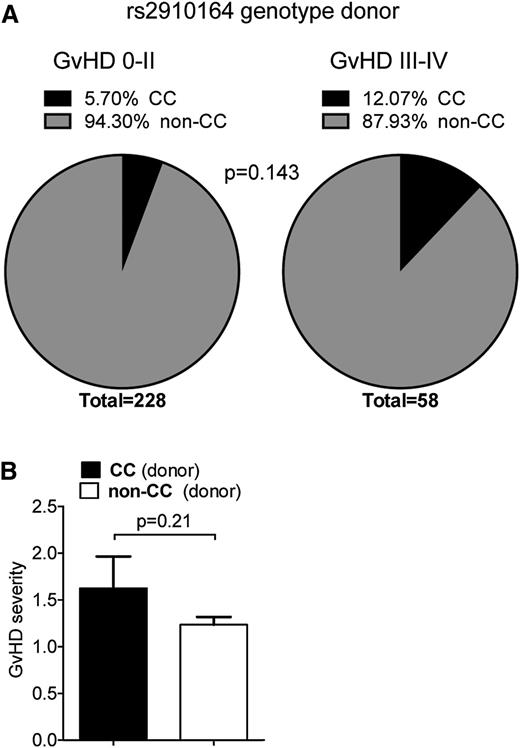
To evaluate the potential relevance of the mouse findings to the human disease, we performed an SNP analysis of genomic DNA from allo-HCT recipients and donors. The SNP rs2910164 causes a G to C base change on genomic DNA. This leads to a mispairing in the hairpin sequence of the precursor of miR-146a, which affects processing of the miRNA precursor and consequently lowers expression of the mature sequence.26 Because the CC genotype, where both alleles carry the SNP, has previously been shown to be linked with higher susceptibility to Crohn's disease,27 we investigated if this SNP might be associated with GVHD severity. Therefore, we analyzed the rs2910164 genotype of 286 hematopoietic cell donors. The distribution of GVHD risk factors in patients with a non-CC or CC genotype donor was equal (Table 1). The observed genotype frequencies for all donors were in accordance with Hardy-Weinberg equilibrium (P > .6). We found that the presence of the CC genotype in HCT donors conferred a trend toward a higher risk for severe GVHD (grade III-IV) in allo-HCT recipients (odds ratio = 2.27, 95% confidence interval = 0.86-5.98, P = .143; Figure 6A; supplemental Table 1): 5.70% of the donors of patients with no or only mild GVHD were homozygous for the risk allele (C), which is comparable to frequencies reported in a large European multicenter study.28 In contrast, >12% of the HCT donors of patients with severe GVHD carried the CC genotype. Likewise, we found a trend toward overall higher GVHD severity in patients when the HCT donor carried the CC genotype (Figure 6B).
miR-146a in human GVHD. (A-B) The rs2910164 SNP genotype was analyzed in 286 HCT donors using Taqman realtime PCR assays. (A) Frequency of donors with the CC genotype vs the non-CC genotype in the group of patients with no/mild GVHD (grade 0-II) and severe GVHD (grade III-IV), respectively, is shown. Frequency of the CC genotype was higher in HCT donors of patients developing severe GVHD compared with no/mild GVHD. (B) Mean GVHD severity in recipients that have received hematopoietic cells from allogeneic donors with the CC genotype vs without the CC genotype is shown.
miR-146a in human GVHD. (A-B) The rs2910164 SNP genotype was analyzed in 286 HCT donors using Taqman realtime PCR assays. (A) Frequency of donors with the CC genotype vs the non-CC genotype in the group of patients with no/mild GVHD (grade 0-II) and severe GVHD (grade III-IV), respectively, is shown. Frequency of the CC genotype was higher in HCT donors of patients developing severe GVHD compared with no/mild GVHD. (B) Mean GVHD severity in recipients that have received hematopoietic cells from allogeneic donors with the CC genotype vs without the CC genotype is shown.
These findings support the concept that miR-146a plays a role in human GVHD pathophysiology, most likely as a rescue mechanism to counteract uncontrolled inflammation.
Discussion
The immune system's response to pathogens and injury must be tightly regulated with regard to its onset and termination. Recognition of the role of miRNAs as central regulators controlling a variety of biological processes at the level of post-transcriptional repression in different inflammatory conditions has increased in recent years. However, miRNA studies in the field of GVHD are still at an early stage, and a recent review highlighted the importance of exploring the role of miRNAs in this disease.29 Using both gain- and loss-of-function approaches, we show that miR-146a negatively regulates GVHD severity and mortality. Consistent with an anti-inflammatory role for miR-146a in GVHD, 2 hallmarks of the disease, proinflammatory cytokine production and alloantigen driven T-cell expansion,30 were both increased when miR-146a−/− T cells were transferred or studied in vitro compared with miR-146a+/+ T cells.
Analyzing a variety of metrics related to T-cell phenotype and function, we observed increased levels of TNF in miR-146a–deficient T cells compared with WT T cells at the RNA and protein levels when stimulated with alloantigen, whereas there was no difference in other T cell-derived cytokines, including IL-4, IL-6, IFN-γ, and IL-17A. Because TNF blockade could prevent the detrimental effects of miR-146a deficiency in T cells and miR-146a overexpression had no effect on Tnf-deficient T cells in terms of their alloreactivity, we propose that miR-146a negatively regulates alloreactive T cells by decreasing their production of TNF.
The role of TNF in acute GVHD has been previously shown to be mediated by direct toxic effects on target tissues and promotion of inflammation.31 Although it had long been presumed that most of the TNF responsible for GVHD came from host or donor monocytes/macrophages, earlier studies have shown that donor T cell-derived TNF is required for GVHD activity.23,24 Our findings are also compatible with in vitro studies showing that miR-146a promotes the binding of the transcription repressor RelB to the TNF promoter, thereby reducing TNF transcription.32
Moreover, in mice, TNF down-modulates the function of Tregs,33 which protect against GVHD.14,34 Although a role for miR-146a in Treg function to control Th1 responses has been described,15 we did not find any numeric or functional differences in Tregs during an allogeneic in vitro or in vivo response (supplemental Figure 2E,H; data not shown).
In humans, elevated TNF levels in the serum35 and intestinal tract36 of patients developing acute GVHD have been described. TNF inhibition using etanercept led to reduced GVHD rates37 ; however, the overall benefit from TNF inhibition in GVHD patients was limited. Because diet-based induction of miR-146a by DIM ameliorated GVHD, it is conceivable that the intestinal immune system may benefit from miR-146a induction. In contrast to TNF inhibition by etanercept in vivo, which can reduce the biological activity of already secreted TNF and is always partial, a miR146a mimic would act further upstream and prevent the secretion of TNF. A miR-146a mimic could therefore interfere earlier with the TNF activity than the inhibitor and prevent autocrine TNF feedforward signaling more efficiently, which could be a potential advantage in the prevention of GVHD. The impact of a miR-146a mimic on the capability of T cells to recognize tumors or pathogens needs to be investigated in further studies.
The unbiased gene expression analysis helped us to identify genes that are differentially regulated in miR-146a−/− T cells compared with miR-146a+/+ T cells following alloantigen stimulation. MiR-146a has been predicted to base pair with sequences in the 3′ untranslated region of the Traf6 transcript,8 and Traf6 has been described as a miR-146a target in several studies.3,9,38,39 Consistent with a role for TRAF6 in alloantigen stimulated T cells, it was one of the most significantly up-regulated genes in miR-146a−/− T cells compared with miR-146a+/+ T cells. In the T-cell receptor signaling pathway that leads to NF-κB activation, TRAF6 mediates signal transduction from the CARMA-1/Bcl-10/MALT1 signalosome to IKK via polyubiquitination of IKKγ.25 Activated IKK in turn phosphorylates the inhibitor of NF-κB (IκB), leading to its ubiquitination and subsequent degradation. In agreement with this model, we found decreased IκB levels in allogeneically stimulated miR-146a−/− T cells. IκB degradation releases NF-κB, enabling its translocation to the nucleus where it activates transcription of target genes, including Tnf. Therefore, the expression of TRAF6 is closely linked with Tnf transcription and, based on our findings, we propose that loss of TRAF6 repression in miR-146a−/− T cells increases IκB degradation, resulting in the observed increase in TNF levels (Figure 5F).
In humans, we found that allo-HCT donors who were homozygous for a miR-146a impairing SNP conferred a trend toward a higher risk for severe GVHD grades III and IV in allo-HCT recipients. Consistent with this finding, an anti-inflammatory role for miR-146a was shown by several SNP studies in which SNPs that lower miR-146a expression were associated with disease activity in Crohn's disease,27 lupus,28 and asthma.40 Larger studies are needed to confirm our results because of the limited number of donors analyzed.
In summary, we report a novel anti-inflammatory role for miR-146a in acute GVHD. Mechanistically, we propose that miR-146a inhibits the TRAF6/NF-κB axis, leading to reduced TNF levels in donor T cells and diminished GVHD severity. Because GVHD severity was decreased when miR-146a expression was enhanced by miRNA mimics or DIM treatment, and because we observed a role for miR-146a in patients developing GVHD, the therapeutic augmentation of miR-146a represents a novel targeted approach to reduce the mortality rate of this severe complication of allo-HCT.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft, Germany (Heisenberg Professorship ZE 872/3-1 to R.Z., DFG individual grant DFG ZE 872/1-2 to R.Z.), in part by SFB620 project C7 to U.S., and the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School to N.S. and R.Z. and Centre for Biological Signaling Studies (BIOSS) II, project B13 to R.Z.).
Authorship
Contribution: N.S. designed the experiments, performed the experiments, evaluated the data and helped to write the manuscript; G.P. helped with experiments; D.P. performed microarray analysis; M.F. helped with qPCR; P.H. helped with patient data collection; A.S.-G. performed histopathological scoring; R.T. helped to design the clinical study; J.F. helped to analyze the patient samples and to write the manuscript; U.S. helped to design and perform the SNP genotype analysis; J.D. helped to develop the concept and analyze the data; and R.Z. designed the overall concept, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert Zeiser, Department of Hematology and Oncology, Freiburg University Medical Center, Albert Ludwigs University, Hugstetterstr. 55, 79106 Freiburg, Germany; e-mail: robert.zeiser@uniklinik-freiburg.de.