Key Points
High-resolution matching for HLA-A, -B, -C, and -DRB1 is required for optimal survival in myeloablative-unrelated donor transplantation.
HLA-DPB1 nonpermissive mismatches should be avoided in otherwise matched transplants to minimize overall mortality.
Abstract
We examined current outcomes of unrelated donor allogeneic hematopoietic cell transplantation (HCT) to determine the clinical implications of donor-recipient HLA matching. Adult and pediatric patients who had first undergone myeloablative-unrelated bone marrow or peripheral blood HCT for acute myelogenous leukemia, acute lymphoblastic leukemia, chronic myelogenous leukemia, and myelodysplastic syndrome between 1999 and 2011 were included. All had high-resolution typing for HLA-A, -B, -C, and -DRB1. Of the total (n = 8003), cases were 8/8 (n = 5449), 7/8 (n = 2071), or 6/8 (n = 483) matched. HLA mismatch (6-7/8) conferred significantly increased risk for grades II to IV and III to IV acute graft vs host disease (GVHD), chronic GVHD, transplant-related mortality (TRM), and overall mortality compared with HLA-matched cases (8/8). Type (allele/antigen) and locus (HLA-A, -B, -C, and -DRB1) of mismatch were not associated with overall mortality. Among 8/8 matched cases, HLA-DPB1 and -DQB1 mismatch resulted in increased acute GVHD, and HLA-DPB1 mismatch had decreased relapse. Nonpermissive HLA-DPB1 allele mismatch was associated with higher TRM compared with permissive HLA-DPB1 mismatch or HLA-DPB1 match and increased overall mortality compared with permissive HLA-DPB1 mismatch in 8/8 (and 10/10) matched cases. Full matching at HLA-A, -B, -C, and -DRB1 is required for optimal unrelated donor HCT survival, and avoidance of nonpermissive HLA-DPB1 mismatches in otherwise HLA-matched pairs is indicated.
Introduction
Allogeneic hematopoietic cell transplantation (HCT) from unrelated donors can be a curative therapy for hematologic malignancies and other blood disorders. Optimizing the outcome of unrelated donor transplantation is vitally important, as the majority of HCT-eligible patients will not have a matched sibling donor. Previous studies demonstrated the adverse impact of donor-recipient HLA mismatch on HCT outcomes1-10 ; however, further progress is needed: although 8/8 (donor-recipient match at HLA-A, -B, -C, and -DRB1) matching results in superior survival,3 there is still substantial graft versus host disease (GVHD) and mortality after matched HCT. As well, selection of the optimal 7/8 matched donor remains a challenge. The importance of HLA-DPB1 and -DQB1 typing remains uncertain and is not routinely performed due to cost and lack of an association with survival in some previous studies. However, recent analyses suggest that HLA-DPB1 classification according to T-cell epitope grouping can identify permissive and nonpermissive donor-recipient combinations relevant to severe acute GVHD (aGVHD) and mortality.11 Validation of these findings could support inclusion of HLA-DPB1 typing and functional classification in routine initial HLA typing. Finally, the relevance of prior analyses has been questioned, as there have been changes in HCT technology and application over time. Most notably, these have included decline in HCT for chronic myelogenous leukemia, utilization of myeloablative conditioning regimens that do not contain total body irradiation, predominant use of peripheral blood stem cells over bone marrow, and improvements in supportive care. Such changes may contribute to observed improvement in survival after HCT over time12 and alter the effects of HLA mismatching on outcomes. To address these issues, we performed a contemporary analysis of HLA matching and unrelated donor HCT outcome.
Methods
Study population
All research was conducted with the approval of the National Marrow Donor Program (NMDP) Institutional Review Board. Unrelated donor transplants were facilitated by the NMDP, and outcomes were reported to the Center for International Blood and Marrow Transplant Research (CIBMTR), NMDP’s research program conducted in collaboration with the Medical College of Wisconsin. Included patients were adults or children with diagnoses of acute myelogenous leukemia (AML), acute lymphoblastic leukemia (ALL), chronic myelogenous leukemia (CML), and myelodysplastic syndrome (MDS), who underwent first myeloablative-unrelated bone marrow or peripheral blood stem cell transplantation conducted between 1999 and 2011. From the total 8003 donor-recipient pairs, 4547 (57%) had HLA typing at HLA-A, -B, -C, and -DRB1 performed through the NMDP retrospective high-resolution typing project. The remaining HLA typing (n = 3456; 43%) was reported to the CIBMTR by the transplant centers and validated by the NMDP. HLA typing information for HLA-DQB1 and -DPB1 was available for 96% and 63% of cases, respectively. Overall survival did not differ between those with HLA-DPB1 typing available vs those without. Myeloablative conditioning was defined by the following: single-dose total body irradiation (TBI) >500 cGy or >800 cGy total in fractionated doses, busulfan of ≥9 mg/kg, melphalan with dose >150 mg/m2, or thiotepa dose >10 mg/kg. Early-stage disease included AML and ALL in first complete remission, CML in first chronic phase, and MDS subtype refractory anemia. Intermediate-stage disease included AML or ALL in second or subsequent complete remission or in first relapse or CML in accelerated phase or second chronic phase. Advanced-stage disease included AML in second or greater relapse or primary induction failure, CML in blast phase, MDS subtype refractory anemia with excess blasts or in transformation, or MDS not otherwise classified. Patients who received lower-intensity conditioning therapy (n = 4691) did not have HCT between 1999 and 2011 (n = 4425), had <6/8 matched HCT (n = 177), did not provide consent for analysis of clinical data (n = 194), were alive with <100 days of follow-up (n = 49), or had missing data on key inclusion criteria (n = 3113) were excluded. All recipients included in this analysis provided informed consent for participation in the NMDP research program, in accordance with the Declaration of Helsinki. A modeling process was used, as previously described,3,13 to adjust for any bias introduced by the exclusion of nonconsenting survivors. This adjustment is standard for all studies using NMDP data. From all potential follow-up data (person-time), 99% was available at 1 year and 92% at 5 years.
HLA typing
High-resolution typing was performed as previously described for HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1.14 Low-resolution (serologic or antigen-level) disparities were derived through conversion of DNA-based typing to serologic equivalent according to the 2010 World Health Organization Nomenclature for factors of the HLA system.15 Mismatch at HLA-DQ (and -DP) included only HLA-DQB1 (and HLA-DPB1), as there is strong linkage disequilibrium between the α and β subunits (>98%), and HLA-DQA1 and HLA-DPA1 typing data are limited (not available in 68% and 80%, respectively, of cases in our dataset). As previously described, we considered the directionality of mismatch for the analysis of GVHD and engraftment.16 Mismatches at homozygous alleles were considered single mismatches. Donor-recipient high-resolution HLA matching at HLA-A, -B, -C, and -DRB1 defined an 8/8 matched pair. Allele- or antigen-level mismatch at 1 (7/8) or 2 (6/8) of these loci defined mismatch groups of interest in the main analysis. We excluded cases (n = 177) that had <6/8 matching, as this practice is infrequent and has prohibitively high mortality. Secondary analyses examined the following: mismatch at HLA-DPB1 or -DQB1, HLA-C*03:03/03:04 vs other -C allele or antigen mismatch,17 and HLA-DPB1 permissive vs nonpermissive mismatches according to T-cell epitope grouping, as previously reported.11,18,19 An online calculator is also available (http://www.ebi.ac.uk/ipd/imgt/hla/dpb.html). The HLA-DPB1 permissive mismatch analysis was performed in both 8/8 and separately in 7/8 cases. These analyses did not consider HLA-DQB1, as HLA-DQB1 mismatch was infrequent (allele matched in 87% of cases), and did not affect survival in our analysis.
Outcome definitions
Overall mortality was defined as time from HCT to death from any cause. Treatment failure was defined as time from HCT to death or primary malignancy relapse. Treatment-related mortality (TRM) was death in continuous remission from the primary malignancy. Relapse was defined per CIBMTR criteria.1 Grades II to IV and III to IV aGVHD were defined by the Glucksberg scale,20 and chronic GVHD was defined as limited or extensive chronic GVHD according to the Seattle criteria.21
Statistical methods
Descriptive statistics included medians and ranges for continuous variables and frequencies for categorical variables. Death was considered a competing risk event for all outcomes except overall mortality and treatment failure, and relapse was considered a competing risk event for estimation of TRM. Patients were censored at time of second HCT or if alive at last follow-up.
The association of number and type of HLA mismatches and clinical outcomes was studied using multivariate proportional hazards models. Mismatched pairs were compared with HLA-matched pairs, allowing precise estimates of the association of mismatch of certain number (1 or 2), type (antigen or allele), and locus (HLA-A, -B, -C, -DRB1, -DPB1, or -DQB1). P < .01 was considered significant. Models were tested for additional significant covariates including patient age, recipient gender, race, Karnofsky performance status (KPS) at HCT, disease, disease stage, time from diagnosis to HCT, graft type (bone marrow vs peripheral blood), donor age, donor parity, donor/recipient gender match, donor/recipient cytomegalovirus (CMV) serostatus match, donor/recipient ABO match, conditioning therapy (TBI-based vs not), GVHD prophylaxis, T-cell depletion (separately considered ex vivo T-cell depletion and in vivo T-cell depletion including ATG and campath), and year of HCT. Models included clinical factors related to the studied outcome at P < .01. All variables were tested for affirmation of the proportional hazards assumption and to investigate interactions with HLA matching. If the proportional hazards assumption was not satisfied, the variable was included as a time-dependent covariate in the model. In the analysis of overall mortality and treatment failure, interaction was detected between disease stage and HLA matching. Thus, the effect of HLA match on overall mortality and treatment failure was performed separately for early-, intermediate-, and advanced-stage disease. No other significant interactions were detected.
The primary analysis (n = 8003) tested the impact of allele or antigen-level mismatch at HLA-A, -B, -C, and -DRB1 on clinical outcomes, comparing 7/8 or 6/8 pairs to 8/8 matched pairs. These findings were validated in a separate analysis only considering unique cases (n = 5846) that did not overlap with previous studies.3,22 Additional analyses tested the effect of mismatch at individual HLA loci at HLA-A, -B, -C, and -DRB1 and allele vs antigen-level mismatch. The impact of allele- or antigen-level mismatch at HLA-DQB1 (n = 7716) and -DPB1 (n = 5015) was examined in separate models for otherwise 8/8 or 7/8 matched pairs. The impact of HLA-C*03:03/03:04 was addressed in a separate analysis comparing those with HLA-C*03:03/03:04, other C allele mismatches, C antigen-level mismatch, other mismatches, and 8/8 matched pairs. Finally, an analysis of those with HLA-DPB1 typing available compared permissive, nonpermissive, and fully matched HLA-DPB1 groups in 8/8 and separately 7/8 matched pairs.11 These findings were confirmed in analyses only considering cases that did not overlap with previous reports (n = 2738).11 As a secondary approach for comparison with prior analyses, we repeated the HLA-DPB1 permissive mismatch analysis in 10/10 and 9/10 cases.
Results
Patient characteristics
Of the study population (n = 8003), cases were 8/8 (n = 5449), 7/8 (n = 2071), or 6/8 (n = 483) matched. Full patient characteristics are presented in Table 1. Median follow-up for surviving patients was 49 (range, 3-151) months. The study population was 88% white, 67% had a KPS of 90 to 100, 77% had acute leukemia, and only 14% had CML. In contrast to prior studies, 56% received a peripheral blood stem cell graft, and 43% had non–TBI-based conditioning. The majority received calcineurin inhibitor-based GVHD prophylaxis. There were no significant differences in pharmacologic GVHD prophylaxis, in vivo, or ex vivo T-cell depletion across locus of HLA mismatch among single mismatch (7/8) cases. Other non-HLA variables differed among 8/8 vs 7/8 vs 6/8 groups: mismatched donors were more often used for younger and nonwhite recipients, ALL or CML diagnoses, intermediate/advanced disease status, at later time from diagnosis to HCT, and using bone marrow vs peripheral blood. HCT therapy among mismatched cases had greater TBI-based conditioning and use of in vivo and ex vivo T-cell depletion. Utilization of 6/8 donors decreased over the studied time period.
Effect of mismatch at HLA-A, -B, -C, and -DRB1
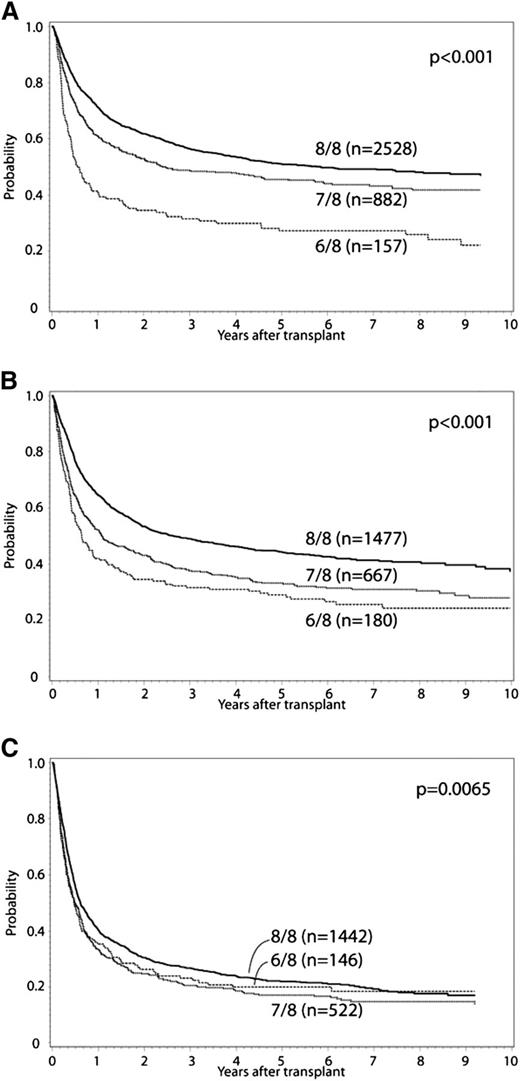
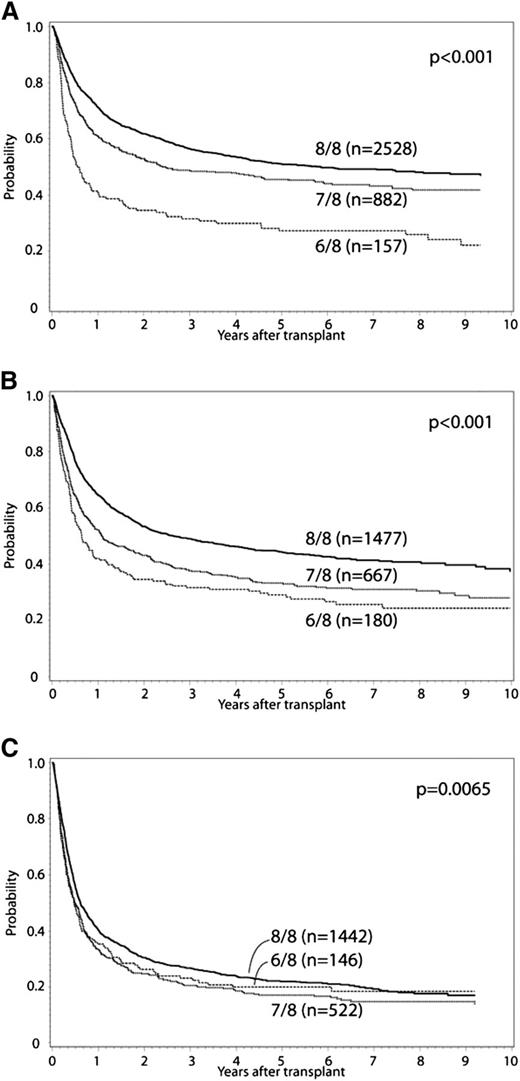
In the primary multivariate analysis, the effect of single (7/8) or double (6/8) locus (HLA-A, -B, -C, or -DRB1) donor-recipient mismatch was examined (Tables 2 and 3). HLA mismatch (6-7/8 vs 8/8) conferred significantly increased risk for grade II to IV and III to IV aGVHD, chronic GVHD, TRM, treatment failure, and overall mortality. Mismatched transplants had a greater proportion of deaths due to GVHD, infection, and organ failure compared with 8/8 matched cases. Malignancy relapse was not affected by HLA mismatch at HLA-A, -B, -C, or -DRB1. These effects were confirmed in separate multivariate models that did not include cases reported in previous analyses.3,22 Additional variables significantly associated with overall mortality included graft type, year of HCT, recipient age, race, KPS, disease, donor-recipient CMV matching, donor-recipient ABO minor and major mismatch, use of TBI, and GVHD prophylaxis (Table 3). Older donor age was associated with increased TRM, but was not significantly associated with overall mortality. There was no significant interaction between year of HCT and the main effect, and analysis results did not differ when restricted to HCT in 2007 to 2011. There was significant interaction between the main effect (HLA mismatch) and disease status for the outcomes of treatment failure and overall mortality. Accordingly, separate analyses were conducted for early-, intermediate-, and advanced-stage disease. The adverse impact of HLA mismatch was greatest among those with early or intermediate stage disease. Survival for early-, intermediate-, and advanced-stage disease patients according to degree of HLA mismatch is presented in Figure 1; adjusted survival curves represent multivariate modeled data. In comparison with 6/8 cases, 7/8 cases had significantly decreased risk for TRM (relative risk [RR], 0.8; 95% confidence interval [CI], 0.69-0.93; P = .003), and among early-stage disease improved treatment failure (RR, 0.65; 95% CI, 0.53-0.8; P < .0001) and overall mortality (RR, 0.62; 95% CI, 0.5-0.76; P < .0001). Significant differences between 7/8 and 6/8 cases were not detected for grade II to IV and III to IV aGVHD or chronic GVHD.
Adjusted OS curves stratified for 8/8, 7/8, and 6/8 separately. (A) early, (B) intermediate, and (C) advanced disease.
Adjusted OS curves stratified for 8/8, 7/8, and 6/8 separately. (A) early, (B) intermediate, and (C) advanced disease.
HLA locus-specific effects
As presented in Table 4, comparably increased risk for aGVHD, chronic GVHD, TRM, treatment failure, and overall mortality was observed for each individual mismatched HLA locus among 7/8 cases. None were significantly associated with risk for relapse. Although the RR was generally lower for mismatch at the -DRB1 locus, power was limited in this subgroup due to sample size. Direct pairwise comparisons between mismatched loci revealed the following: single mismatch at -B had greater risk for grade III to IV aGVHD (RR, 1.4; 95% CI, 1.1-1.9; P = .008), chronic GVHD (RR, 1.3; 95% CI, 1.1-1.6; P = .003), and lower risk for relapse (RR, 0.65; 95% CI, 0.5-0.85; P = .0015) compared with single mismatch at -C. No other significant differences in outcomes were observed between mismatched loci.
Comparison of allele vs antigen-level mismatch
Both allele and antigen-level mismatches were associated with increased risk for grade III to IV aGVHD, TRM, treatment failure, and overall mortality. There was no association with relapse. Significant differences between allele and antigen-level mismatch in aggregate (not considering individual mismatch loci) were not detected for any of the studied outcomes. Allele vs antigen-level comparison at each individual HLA locus supported that B allele mismatch had decreased risk for grade II to IV aGVHD compared with B antigen mismatch (RR, 0.56; 95% CI, 0.4-0.78; P = .0007). No other differences were detected between allele vs antigen level mismatch in the locus-specific analysis. We found no statistically significant difference between HLA-C*03:03/03:04 and other HLA-C allele mismatches, but had limited power due to small sample size in each subgroup. However, consistent with a recent study,17 we observed an increased risk for overall mortality between the other HLA-C allele mismatched cases and 8/8 matched group, but not for the pairs with HLA-C*03:03/03:04. Similar results were observed when the analysis was restricted to nonoverlapping subjects.
Effect of HLA-DQB1 and HLA-DPB1 mismatch
Among 8/8 matched cases, HLA-DQB1 mismatch was associated with grade II to IV aGVHD (single allele mismatch: RR, 1.2; 95% CI, 0.96-1.5; P = .1; single antigen mismatch: RR, 1.4; 95% CI, 1.1-1.7; P = .006). No significant difference was observed between single allele and single antigen HLA-DQB1 mismatch. DQB1 mismatch was not associated with other studied outcomes. Among 7/8 matched cases, no significant effects of HLA-DQB1 mismatch were observed.
Among 8/8 matched cases, HLA-DPB1 mismatch was associated with increased risk for grade II to IV (single allele mismatch: RR, 1.4; 95% CI, 1.2-1.6; P = .002; double allele mismatch: RR, 1.6; 95% CI, 1.3-1.9; P < .0001), grade III to IV aGVHD (single allele: RR, 1.5; 95% CI, 1.1-2.0; P = .004; double allele mismatch: RR, 1.7; 95% CI, 1.3-2.3; P = .0004), and decreased relapse (single allele mismatch: RR, 0.71; 95% CI, 0.6-0.8; P < .0001; double allele mismatch: RR, 0.7; 95% CI, 0.6-0.85; P = .0002) compared with HLA-DPB1 allele matched cases. No significant differences were observed between single and double allele HLA-DPB1 mismatches. DPB1 mismatch was not associated with risk for other studied outcomes. Among 7/8 matched cases, no significant effects of HLA-DPB1 mismatch were observed, but numbers of evaluable cases were substantially less.
T-cell epitope matching-based HLA-DPB1 classification
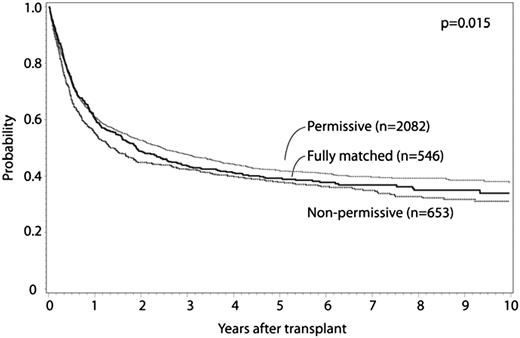
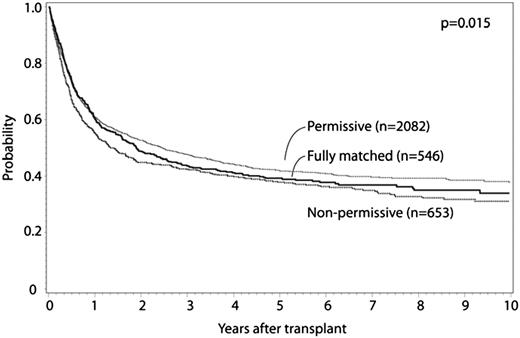
HLA-DPB1 allele mismatches were categorized for 8/8 matched cases as previously described.11 Fully HLA-DPB1 allele matched and nonpermissive HLA-DPB1 allele mismatched cases were compared with permissive HLA-DPB1 allele mismatched cases. Patient characteristics are presented in Table 5, multivariate analysis results are presented in Table 6, and survival curves are presented in Figure 2; adjusted survival curves represent multivariate modeled data. Both permissive and nonpermissive HLA-DPB1 allele mismatches were associated with increased risk for grade II to IV and grade III to IV aGVHD compared with matched cases, with no significant difference in risk in the comparison of permissive and nonpermissive cases. Similarly, both permissive and nonpermissive HLA-DPB1 allele mismatches were associated with significant decrease in relapse risk compared with HLA-DPB1 allele matched cases, with no difference in relapse observed between permissive and nonpermissive cases. Nonpermissive HLA-DPB1 allele mismatched cases had significantly greater TRM compared with either permissive HLA-DPB1 allele mismatched or HLA-DPB1 allele matched cases. Nonpermissive HLA-DPB1 allele mismatch was associated with significantly greater overall mortality compared with permissive HLA-DPB1 allele mismatch, although no significant differences were detected for permissive or nonpermissive cases compared with matched. No significant interaction was found between disease status and HLA-DPB1 mismatch categories. No significant effect of other donor characteristics (eg, age, CMV, and ABO matching) was observed in this model. These findings were confirmed in separate multivariate analyses that excluded cases shared with a previously published study.11 No significant differences in outcome were observed according to single vs double HLA-DPB1 allele mismatches among permissive and nonpermissive cases. No significant differences in outcomes were observed between HLA-DPB1 permissive and nonpermissive mismatches when the analysis was limited to 7/8 cases, although evaluable patients were substantially less compared with the 8/8 analysis. In a separate analysis, similar conclusions were reached when considering 10/10 matched cases (Table 7). No significant differences were observed when limited to 9/10 matched cases.
Adjusted OS curves for -DPB1 matched, permissive mismatch, and nonpermissive mismatch cases.
Adjusted OS curves for -DPB1 matched, permissive mismatch, and nonpermissive mismatch cases.
Discussion
In the analysis of a recent transplant population representative of changes in HCT technology, we demonstrate the importance of high-resolution typing of unrelated donors and recipients for HLA-A, -B, -C, and -DRB1 and confirm that single allele or antigen-level mismatch (7/8 match) is associated with increased risk for severe aGVHD, chronic GVHD, TRM, and greater overall mortality. The effect on overall mortality is most apparent among those with early stage disease. Although 7/8 matched donors should be considered an acceptable option among those without 8/8 matched donors, 6/8 matched transplants are associated with prohibitively high TRM and should not routinely be used. Although high-quality comparative data are not available, such patients may derive greater benefit from other approaches or other stem cell sources, such as umbilical cord blood or haploidentical transplantation.23 In contrast to some previous reports, the data do not consistently support that allele vs antigen or specific mismatched loci significantly alter outcomes. Few differences were observed between B antigen and B allele (grade II to IV aGVHD), as well as B vs C mismatch (grade III to IV aGVHD, chronic GVHD, and relapse); however, none of these altered overall mortality. It should be noted, however, that our study of HLA-C allele mismatches did support a comparable HR for overall mortality for HLA-C*03:03/03:04 and 8/8 matches, in keeping with a recently reported analysis.17
The data confirm the adverse impact of nonpermissive HLA-DPB1 mismatch on TRM and overall mortality in an independent data set (excluding overlap from a previous analysis of Fleischhauer et al) among 8/8 matched pairs and 10/10 matched pairs.11 Validation of these findings is particularly noteworthy, given other major differences in study populations. In that prior analysis, 47% of cases were from the Japan Marrow Donor Program, 90% used bone marrow rather than peripheral blood stem cells, and 68% received TBI-based myeloablative conditioning.11 In contrast to this prior report, we found that any HLA-DPB1 mismatch was associated with increased aGVHD but could not identify differences between permissive and nonpermissive mismatch for this outcome. We found that nonpermissive DP mismatched cases had increased risk for aGVHD compared with matched cases, regardless of the mismatch vector (GVH vs host versus graft vector). Previous data suggested similar effects, and investigators have proposed mechanistic hypotheses for this finding.11,18,19 In aggregate, the data suggest that donor selection to avoid nonpermissive HLA-DPB1 mismatches may result in improved survival after unrelated donor HCT in pairs that are otherwise 8/8 or 10/10 matched. We acknowledge that -DPB1 permissive mismatches had both advantages (reduced malignancy relapse and improved treatment failure) and risks (increased risk for grade II to IV and III to IV aGVHD) compared with HLA-DPB1 allele matched pairs. These differences may support selection of HLA-DPB1 permissive mismatched or HLA-DPB1 allele matched donors in individual circumstances. However, based on the lack of overall mortality difference between these options, our major recommendation is to avoid nonpermissive HLA-DPB1 mismatches. Our data do not support an impact of nonpermissive HLA-DPB1 mismatch among 7/8 or 9/10 matches in contrast with the prior report,11 which may be explained by lower sample size in the permissive and nonpermissive HLA-DPB1 groups in our study. It is estimated that nonpermissive HLA-DPB1 alleles occur in ∼30% of the population,11,24 and a recent study suggested, among those with otherwise comparable donor options, consideration of HLA-DPB1 types may permit skewing toward donors with permissive mismatch.25 An algorithm for calculating the number of donors required to achieve a permissive DPB1 mismatch, according to the T-cell epitope (TCE) group of the patient, has been developed; this builds on observed HLA-DPB1 allele frequencies in a prior study.26 For patients belonging to TCE groups 1, 2, and 3 respectively, the total number of donors typed to achieve 50% probability of a permissive DPB1 mismatch is estimated to be 9, 4, and 1. The corresponding figures for 90% probability are 29, 11, and 2. More than 95% of subjects in this prior analysis belonged to TCE groups 2 and 3.26 Thus, these estimates support that permissive DPB1 mismatch could be identified for the majority of patients with a modest number of donors typed. A prospective study focused on validation of the projected TCE permissive match rates is currently underway at the NMDP. Incorporation of HLA-DPB1 typing in unrelated donor searches varies, and implementation of this strategy must consider additional time and cost incurred, as well as urgency of donor selection and transplantation in individual patient scenarios.
In keeping with some,27 but not other,1,3 prior investigations, we identified HLA-DQB1 mismatch as a risk factor for grade II to IV aGVHD. This effect was most apparent among cases with single antigen mismatch, although we could not specifically identify a differential effect according to allele vs antigen-level mismatch at HLA-DQB1. As well, these effects were only detected among 8/8 cases and not in the setting of 7/8 match. In keeping with previous studies, we found no impact of HLA-DQB1 mismatch on mortality.1-3 Thus, selection of HLA-DQB1 matched donors may reduce the risk for aGVHD and could inform rational donor selection when multiple otherwise 8/8 matches are available. Although our analysis does not support HLA-DQB1 mismatch as an independent risk factor for mortality, we acknowledge the previously reported adverse effect of ≥3 low-expression loci mismatches (DRB3/4/5, DQ, and DP) on mortality among otherwise 7/8 matched pairs.28
We note the following limitations to this analysis: First, numbers in certain subgroups limit power to detect differences in outcomes. In particular, the total number of HLA-DRB1 mismatches is relatively low, limiting power for analysis of this locus-specific effect. However, the number of single HLA-DRB1 mismatches in our analysis rivals that of previous studies that demonstrated an effect of HLA-DRB1 mismatch on mortality. Second, we acknowledge that HLA-DPB1 and -DQB1 typing was not available uniformly, and therefore we limited these analyses to only those cases with such data. This reflects the extent of HLA typing performed in current NMDP unrelated donor searches. Third, we used the HLA-DPB1 TCE classification developed by Fleischhauer et al for validation purposes,11 but acknowledge that alternative HLA-DPB1 allele matching algorithms may be warranted. Next, we acknowledge that other non-HLA variables may modify the effect of HLA mismatch on outcome. Accordingly, we accounted for such interactions, specifically investigating early-, intermediate-, and advanced-disease separately. Next, we acknowledge some overlap of our study population with previous analyses. We removed shared cases in secondary analyses and confirmed our primary findings. As well, we have not examined donor-directed HLA-specific alloantibodies in our current study. We acknowledge that prior reports have demonstrated an effect on risk for graft failure29 ; however, our current analysis both had insufficient cases of graft failure for study and no comprehensive data on HLA-specific alloantibodies. Finally, we intentionally limited this analysis to certain diseases and myeloablative conditioning; a separate NMDP/CIBMTR study is examining allied questions in the setting of reduced-intensity conditioning.
The data support that matching at HLA-A, -B, -C, and -DRB1 is required for optimal unrelated donor HCT survival, and avoidance of nonpermissive -DPB1 mismatches in 8/8 or 10/10 matched pairs is indicated. Future work is needed to integrate these findings with previously reported nonpermissive donor-recipient allele combinations and amino acid substitutions to facilitate optimal unrelated donor selection.11,17,28,30-32
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We acknowledge the contribution of B.S. and J.R. in modeling the number of donors required to achieve permissive mismatch according to TCE group.
The Center for International Blood and Marrow Transplant Research (CIBMTR) is supported by Public Health Service grant/cooperative agreement U24-CA076518 from the National Cancer Institute (NCI), the National Heart, Lung and Blood Institute (NHLBI), and the National Institute of Allergy and Infectious Diseases (NIAID), grant/cooperative agreement 5U10HL069294 from the NHLBI and NCI, contract HHSH250201200016C with Health Resources and Services Administration (HRSA/DHHS), grants N00014-13-1-0039 and N00014-14-1-0028 from the Office of Naval Research, and grants from Actinium Pharmaceuticals, Allos Therapeutics, Amgen, an anonymous donation to the Medical College of Wisconsin, Ariad, Be the Match Foundation, Blue Cross and Blue Shield Association, Celgene Corporation, Chimerix, Inc., Fred Hutchinson Cancer Research Center, Fresenius-Biotech North America, Inc., Gamida Cell Teva Joint Venture Ltd., Genentech, Inc., Gentium SpA, Genzyme Corporation, GlaxoSmithKline, Health Research, Roswell Park Cancer Institute, HistoGenetics, Incyte Corporation, Jeff Gordon Children’s Foundation, Kiadis Pharma, The Leukemia and Lymphoma Society, Medac GmbH, The Medical College of Wisconsin, Merck & Co, Millennium: The Takeda Oncology Co., Milliman USA, Miltenyi Biotec, National Marrow Donor Program, Onyx Pharmaceuticals, Optum Healthcare Solutions, Osiris Therapeutics, Otsuka America Pharmaceutical, Perkin Elmer, Remedy Informatics, Sanofi US, Seattle Genetics, Sigma-Tau Pharmaceuticals, Soligenix, St. Baldrick’s Foundation, StemCyte, A Global Cord Blood Therapeutics Co., Stemsoft Software, Swedish Orphan Biovitrum, Tarix Pharmaceuticals, TerumoBCT, Teva Neuroscience, THERAKOS, University of Minnesota, University of Utah, and Wellpoint.
The views expressed in this article do not reflect the official policy or position of the National Institutes of Health, the Department of the Navy, the Department of Defense, Health Resources and Services Administration, or any other agency of the US Government.
Authorship
Contribution: J.P., S.J.L., K.W.A., S.S., and C.A. drafted the research plan; J.P., S.J.L., K.W.A., S.S., C.A., H.-L.W., M. Askar, M. Aljurf, J.D., M.F.V., A.G., V.G., R.H., C.K.H., Y.I., A.A.K., T.N., C.M., M.O., E.W.P., V.P., J.R., W.S., K.R.S., B.S., J.S., W.A.W., and A.E.W. critically revised the research plan; K.W.A. and H.-L.W. performed statistics; J.P., S.J.L., K.W.A., S.S., and C.A. analyzed and interpreted data and drafted the paper; and J.P., S.J.L., K.W.A., S.S., C.A., H.-L.W., M. Askar, M. Aljurf, J.D., M.F.V., A.G., V.G., R.H., M.M.H., C.K.H., Y.I., A.A.K., T.N., C.M., M.O., E.W.P., V.P., J.R., W.S., K.R.S., B.S., J.S., W.A.W., and A.E.W. critically revised the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joseph Pidala, H. Lee Moffitt Cancer Center and Research Institute, Tampa, FL 33612; e-mail joseph.pidala@moffitt.org.