Abstract
Introduction: Peripheral T-cell lymphomas (PTCLs) are a heterogeneous group of non-Hodgkin lymphomas noted for their poor prognosis. Their molecular pathogenesis has not been entirely elucidated. We previously found that primary thyroid T-cell lymphoma (PTTL) is a distinct entity among heterogeneous PTCLs and that this disease is characterized by the genomic loss of 6q24 (Br J Haematol., 161:214-223). In this study, we extended the analysis to other types of PTCLs and performed functional assays to identify causative genes located on 6q24.
Methods: Focusing on chromosome 6q loss, we reexamined previous comparative genomic hybridization data from 267 PTCL cases comprising 6 PTTLs, 51 PTCLs-not otherwise specified (NOS), 62 adult T-cell leukemias/lymphomas, 35 natural killer (NK)-cell lymphomas, 39 angioimmunoblastic T-cell lymphomas (Genes Chromosomes Cancer, 46:37-44), and 74 anaplastic large cell lymphomas (Br J Haematol., 140:516-526). Gene expression levels were determined by using published gene expression profiling (GEP) data (GSE6338 and GSE19069) and quantitative real-time reverse transcription polymerase chain reaction (RT-PCR). Subsequently, we established Tet-Off cell lines belonging to several lineages (6 T-cell lines, 1 NK-cell line, 4 B-cell lines, 1 myeloid cell line, and 3 epithelial cell lines) for functional analyses.
Results: Genomic loss of 6q24 was observed in 8% (n = 267) of PTCL cases, and it occurred most frequently in PTTL cases (67%; n = 6). All the genomic losses were heterozygous; homozygous loss of this region was not observed in our analysis. The smallest region of deletion, observed in a PTTL case, was considered the minimal common region (MCR) of 6q24 loss. The MCR contained 2 known coding genes, STX11 and UTRN. Combined GEP data and quantitative RT-PCR analyses showed that the expression of STX11, but not UTRN, was markedly lower in PTCL than in normal T-cells. We therefore regarded STX11 as the most probable candidate gene located in 6q24. Syntaxin 11, encoded by STX11, is a t-SNARE protein that plays a role in binding vesicles to cell membranes, and alteration of STX11 in the germline causes familial hemophagocytic syndrome type 4.
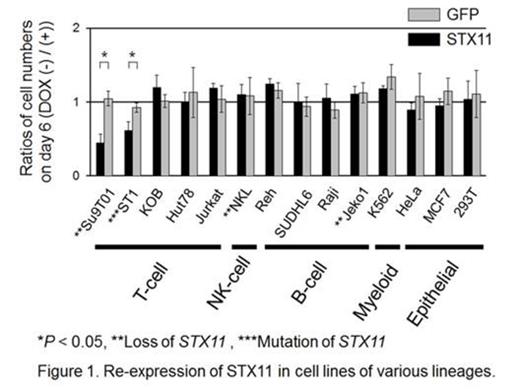
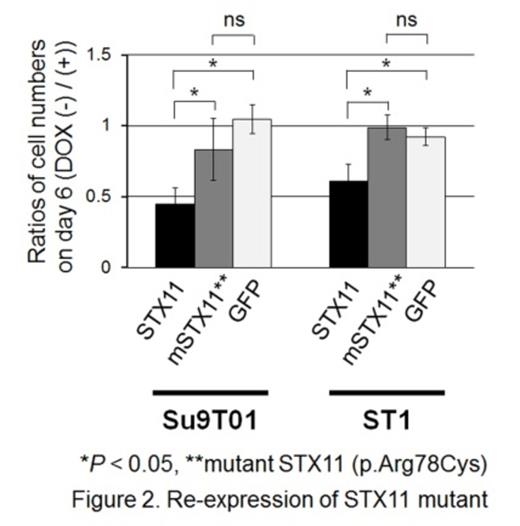
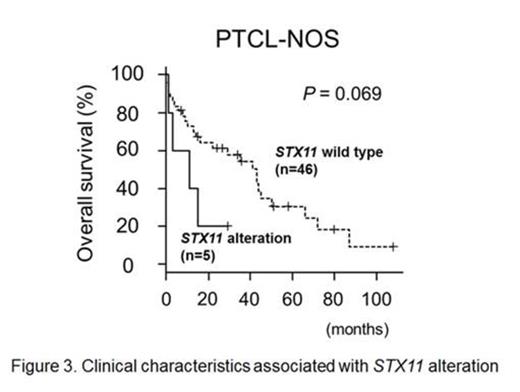
To further evaluate genomic alteration of STX11, mutation analysis was performed on PTCL-NOS and PTTL cases as well as T-cell lines, for which adequate DNA was available. This revealed STX11 mutations in 2 cases (1 PTCL-NOS case and 1 T-cell line). Wild-type STX11 expression suppressed the proliferation of T-cell lines bearing genomic alterations at the STX11 locus only, and it did not show suppressive effects on other lineage cell lines (Fig. 1). Expression of STX11 induced cellular apoptosis in the cell line, although the number of apoptotic cells induced was relatively small. Interestingly, expression of a novel STX11 mutant (p.Arg78Cys), observed in a T-cell line, did not exert suppressive effects on the induced cell lines suggesting that there was a loss-of-function mutation (Fig. 2). Finally, we evaluated the clinical impact of STX11 alteration in PTTL and PTCL-NOS cases where data were available. This showed that PTCL-NOS cases with genomic alterations of STX11 tended to have a poorer prognosis than those without (Fig. 3; P = 0.069).
Conclusion:
In the present study, we examined the MCR of 6q24 loss and showed that STX11 acts as a tumor suppressor gene in PTCLs only. These findings provide a novel approach for understanding the molecular pathogenesis of PTCLs, and they may contribute to the future development of new drugs for the treatment of PTCLs.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.