Abstract
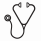
Juvenile Myelomonocytic Leukemia (JMML) is an aggressive myeloproliferative neoplasm of childhood with a 5-year event free survival of 52% after hematopoietic stem cell transplantation (HSCT). A hallmark of JMML is aberrant Ras pathway activation due to mutations in NF1, NRAS, KRAS, PTPN11 and CBL. However, robust predictors of response are lacking, as individual mutations are not reliably associated with outcome, and relapse remains the most common reason for treatment failure. Recently, massively parallel sequencing has identified recurrent mutations in the SKI domain of SETBP1 in a variety of myeloid disorders, including JMML (Piazza et al Nat Genet 2012, Makishima et al Nat Genet 2013, Sakaguchi et al Nat Genet, 2013). These mutations had a lower allelic frequency compared to Ras pathway mutations, but were associated with poor prognosis. These and other data suggested that SETBP1 mutations contribute to disease progression rather than initiation. We identified several patients with JMML who had clonal SETBP1 mutations detected at relapse. Analysis of mononuclear cell extracted DNA from serial samples of two patients who relapsed revealed an increase in the SETBP1 mutant allele frequency over time (Figure 1). Similarly, analysis of colonies plated in methylcellulose from serial time points indicated that the percentage of individual myeloid progenitor colonies that were heterozygous or homozygous for the SETBP1 mutation increased with each sequential sample despite intensive treatment. Based on these data, we tested the hypothesis that rare SETBP1 mutant clones exist at diagnosis in many patients who relapse, and that these rare cells undergo positive selection during treatment. Using a droplet digital PCR (ddPCR) technology with a detection threshold as low as 0.001% of mutant DNA, we identified SETBP1 mutations in 16/53 (30%) of diagnostic JMML specimens from children treated on Children's Oncology Group trial AAML0122. Of these mutations, 12 were subclonal and 4 were clonal. Event free survival (EFS) at 4 years in patients with SETBP1 mutations was 19% ± 10% compared to 51% ± 8% in those with wild type SETBP1 (p=0.006). While samples of patients who relapsed on the AAML0122 trial were not available for analysis, one patient recently undergoing treatment who had a subclonal SETBP1 mutation (0.45% allelic fraction) detected at diagnosis by ddPCR, demonstrated an overt SETBP1 mutation at relapse. Finally, we isolated and analyzed hematopoietic stem (HSC), multipotent progenitor (MPP), common myeloid progenitor (CMP), and granulocyte-monocyte progenitor (GMP) populations from a relapsed sample with a SETBP1 mutation. Sanger sequencing demonstrated that all four progenitor compartments were affected by the mutation. Analysis of additional samples is underway. We conclude that the presence of a subclonal mutation in SETBP1 is a novel biomarker of adverse outcome in JMML. Understanding the mechanisms underpinning SETBP1-mediated resistance and relapse, and further identifying therapeutic vulnerabilities of HSCs expressing these mutant proteins will be critical to improve outcomes for patients with JMML and other myeloid malignancies. Furthermore, the presence of a subclonal SETBP1 mutation at diagnosis might identify JMML patients who will benefit from more intensive conditioning prior to HSCT or from novel therapeutic strategies.
Troup:Bio-Rad Laboratories: Employment.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract