Key Points
Marker genes enable detection and selection of T cells, whereas suicide genes enable selective destruction of T cells in case of toxicity.
RQR8 is a 136-amino-acid epitope-based marker/suicide gene that enables clinical selection, cell tracking, and deletion in case of toxicity.
Abstract
A compact marker/suicide gene that utilizes established clinical-grade reagents and pharmaceuticals would be of considerable practical utility to T-cell cancer gene therapy. Marker genes enable measurement of transduction and allow selection of transduced cells, whereas suicide genes allow selective deletion of administered T cells in the face of toxicity. We have created a highly compact marker/suicide gene for T cells combining target epitopes from both CD34 and CD20 antigens (RQR8). This construct allows selection with the clinically approved CliniMACS CD34 system (Miltenyi). Further, the construct binds the widely used pharmaceutical antibody rituximab, resulting in selective deletion of transgene-expressing cells. We have tested the functionality of RQR8 in vitro and in vivo as well as in combination with T-cell engineering components. We predict that RQR8 will make T-cell gene therapy both safer and cheaper.
Introduction
Adoptive immunotherapy is an established and evolving therapeutic approach. In the setting of allogeneic hematopoietic stem cell transplantation, donor lymphocyte infusions are frequently given to treat relapse of hematologic malignancies. Tumor-infiltrating lymphocytes are effective in treating metastatic melanoma. Genetic engineering of T cells greatly increases the scope and potency of T-cell therapy: T-cell receptor (TCR) transfer allows targeting of intracellular cancer antigens, whereas chimeric antigen receptors (CARs) allow targeting of surface cancer or lineage-specific antigens.1 Clinical responses have been observed with both approaches, and numerous further trials are underway.
Acute adverse events can occur following adoptive immunotherapy. Graft-versus-host disease is a common and serious complication of donor lymphocyte infusions. Administration of engineered T cells has also resulted in toxicity.2-5 For instance, on-target off-tumor toxicity has been reported in native TCR transfer studies against melanoma antigens6 ; T cells redirected to the renal cell carcinoma antigen carbonic anhydrase IX produced unexpected hepatotoxicity.7 Immune activation syndromes have been reported after CD19 CAR therapy.3,8,9 Finally vector-induced insertional mutagenesis results in a theoretical risk of lymphoproliferative disorders. The incidence and severity of these toxicities is unpredictable. Further, in contrast to a therapeutic protein or small molecules, whose adverse events usually abate with the half-life of the therapeutic, T cells engraft and replicate, potentially resulting in escalating and fulminant toxicity.
A suicide gene is a genetically encoded mechanism that allows selective destruction of adoptively transferred T cells in the face of unacceptable toxicity. Two suicide genes have been tested in clinical studies: herpes simplex virus thymidine kinase (HSV-TK)10 and inducible caspase-9 (iCasp9).11 Expression of HSV-TK in T cells confers susceptibility to ganciclovir. HSV-TK is a highly effective suicide gene, but immunogenicity limits application to clinical settings of profound immunosuppression such as haploidentical hematopoietic stem cell transplantation. Further, it precludes the use of ganciclovir for the treatment of cytomegalovirus infection. iCasp9 is activated by an experimental small-molecule dimerizer (AP20187); hence, use of this suicide gene depends on the availability of clinical-grade dimerizer. Finally, both are intracellular proteins and typically must be coexpressed with a marker gene.
Marker genes enable detection and positive selection of transduced T cells. Some T-cell engineering strategies do not result in transgenic expression of readily detectable surface proteins.1 In these cases, measurement of transduction and tracking of cells in peripheral blood is difficult, and a marker gene is useful. Further, in some settings, it is essential to administer only transduced T cells, for instance in graft-versus-host disease suicide-gene protocols. Described marker genes include several truncated type I transmembrane proteins not normally expressed on T-cells: the truncated low-affinity nerve growth factor,12 truncated CD19,11 and truncated CD34.13 A particularly attractive feature of CD19 and CD34 is the availability of the off-the-shelf Miltenyi CliniMACS selection system for clinical-grade sorting. However, these are relatively large surface proteins that may tax the vector packaging capacity and transcriptional efficiency of an integrating vector.
We sought to develop a highly compact epitope-based construct that would act as both a suicide and marker gene. For maximum convenience, we restricted our design to one that could use existing good manufacturing practice reagents and standard pharmaceuticals for both selection and deletion. Our final construct, RQR8, is a 136-amino-acid protein that is recognized by the anti-CD34 antibody QBEnd10, the antibody used in the Miltenyi CliniMACS CD34 selection system. It renders T cells highly susceptible to lysis by the therapeutic monoclonal rituximab. We anticipate RQR8 will make T-cell therapy cheaper, easier, and safer.
Materials and methods
Cloning
All constructs were generated by in-house gene synthesis using polymerase chain reaction assembly of overlapping oligos unless otherwise specified. Codon optimization used an in-house algorithm (written by M.P. and available upon request) that strove to keep GC content at 70% and eliminate cryptic splicing, hairpins, literal repeats, and any possible cis-acting sequences. Identity of constructs was confirmed through capillary sequencing Applied Biosystems 3730xL capillary. Phusion polymerase, quick Ligase, and NEB5α (New England Biolabs) were used for molecular cloning. Oligonucleotides were purchased from IDTDNA. The retroviral vector used in all constructs was the splicing oncoretroviral vector SFG.14 Enhanced green fluorescent protein (eGFP) and enhanced blue fluorescent protein 2 (eBFP2)15 were coexpressed from an encephalomyocarditis virus internal ribosomal entry site sequence.16 Anti-GD2 CAR was as described elsewhere (K.S., T. Gileadi, E.K., B.F., R. Wallace, H. Zhang, N. Westwood, D. Edwards, W. Qasim, J. Anderson and M.P., unpublished data). RQR8 was coexpressed with CAR by cloning it upstream of the CAR separated by an in-frame foot-and-mouth–like 2A peptide, TaV,17 or coexpressed with the wild-type-1–specific TCR α and β chains with 2 interposed foot-and-mouth disease (FMD)-2A TaV sequences, codon wobbled to prevent retroviral recombination. Annotated sequences of constructs used are included in supplemental Data 2 (available at the Blood Web site).
Retroviral production and transduction
RD114-pseudotyped supernatant was generated as follows: 293T cells were transfected with vector plasmid; RDF, an expression plasmid to supply RD114 envelope (gift of Mary Collins, University College London)18 ; and PeqPam-env, a gagpol expression plasmid (gift of Elio Vanin, Baylor College of Medicine).17,19,20 Transfection was facilitated using genejuice (Merck). Eco-pseudotyped supernatant was generated in a similar way, with RDF substituted by the ecotropic envelope expression plasmid pMono.Eco. Peripheral blood mononuclear cell transductions were performed as follows: T cells were isolated by Ficoll (GE Healthcare) gradient centrifugation and stimulated with phytohemagglutinin at 5 µg/mL. Interleukin-2 (IL-2) stimulation (100 IU/mL) was added following overnight stimulation. On day 3, T cells were harvested, plated on retronectin and retroviral supernatant, and centrifuged at 1000g for 40 minutes. Murine splenocytes were isolated and transduced as follows: splenocytes from 6- to 8-week-old C57BL/6 mice were isolated by passage through a 40-µm-pore sieve followed by red blood cell lysis in ACK lysis buffer (Lonza). Splenocytes were activated in RPMI supplemented with 10% fetal calf serum, Glutamax, 10 mM HEPES, and 0.1 µm 2-mercaptoethanol supplemented with 2 µg/mL concanavalin A (Sigma) and 1 ng/mL interleukin-7 (Peprotech) at a density of 1 to 1.5 × 106/mL for 24 hours. Retroviral transduction was performed by loading retronectin-coated plates (Takara) with 2 to 3 × 106 splenocytes suspended in 1 mL retroviral supernatant, centrifuged at 805g for 90 minutes, and then recovered by overnight incubation at 37°C. The following day, splenocytes were recovered from the plate, resuspended in complete RPMI supplemented with 50 ng/mL murine IL-2 (Invitrogen), and cultured overnight. Transduced splenocytes were purified on LS columns using CD34 microbeads (Miltenyi Biotec) as per the manufacturer’s instructions. Following positive selection, splenocytes were resuspended in conditioned media retained from the overnight IL-2 stimulation and cultured overnight. Purity and viability of selected splenocytes were assessed by flow cytometry prior to adoptive transfer 1 day following positive selection.
Antibodies, flow cytometry, and flow sorting
CAR was stained using a Cy5-conjugated polyclonal goat anti-human-Fc (Jackson Immunotech). Rituximab and ofatumumab were procured from the hospital pharmacy at University College London Hospital. Murine immunoglobulin G2a (IgG2a) rituximab was generated by cloning the variable regions of the heavy and light chain from IDEC2B8 hybridoma HB-11388 checked against patent 5 843 439 into the constant regions of murine IgG2a and murine immunoglobulin κ (sourced from InvivoGen plasmids pFUSEss-CH1g-mG2A and pFUSE2ss-CLIg-mk) into a retroviral vector (MP9201 SFG.S-Rtx_H_mIgG2a.I2.eGFP and MP9202 SFG.S-Rtx_L_mKappa.I2.eBFP2; see supplemental Data 2 for sequences). K562 cells were transduced with vectors coding for both heavy and light chains. A highly productive clone was identified, expanded in Phenol Red negative Iscove modified Dulbecco medium supplemented with 2.5% immunoglobulin G–depleted serum (Biosera). Flow cytometry was performed using Beckman-Coulter Cyan and BD LSRII Fortessa instruments. Flow cytometric sorting was performed using a Beckman Coulter MoFlo-XDP sorter.
Complement-mediated cytotoxicity
Transduced T cells were exposed to 25% baby-rabbit complement (AbD Serotec) for 4 hours with or without inclusion of rituximab (100 µg/mL) to examine complement-dependent cytotoxicity (CDC)-mediated sensitivity. Miltenyi CD34 magnetic bead–selected-transduced RQR8 T-cells were compared against a similarly treated population of Q8-transduced T cells to demonstrate specificity of CDC-mediated deletion. Further examination of CDC assay parameters was achieved through time-course/dose-titration assays using RQR8-transduced T cells incubated with rituximab at 12.5, 25, 50, and 100 µg/mL and time-point assessments ranging between 1 to 120 minutes.
Antibody-dependent cellular cytotoxicity
Natural killer (NK) cell effectors were generated using a K562 stimulator cell line (after Campana21 ), expressing membrane-bound interleukin-15 and 41BBL established by retroviral vector transduction and single-cell cloning. Freshly isolated peripheral blood mononuclear cells from healthy donors were cocultured 1:1 in 24-well tissue-culture–treated multiwell plates with irradiated K562.41BBL.mIL15 irradiated at 120 Gy and supplemented with 40iu IL2. Partial media changes performed as required. Following 7 days in culture, a pure population (>95% purity) of NK cells was isolated following a single round of Miltenyi CD56-positive selection. Transduced T-cell targets were cocultured with NK cell effectors at effector:target ratios of 16:1, 8:1, 4:1, and 2:1 for 48 hours. Cellular deletion was assessed by flow cytometry following Annexin V (BD Biosciences) and propidium iodide (PI) (Sigma-Aldrich) staining. For sensitivity assessment of antibody-dependent cellular cytotoxicity (ADCC)-mediated deletion, NK cell effectors were prepared with CellTRACE violet (Invitrogen) immediately prior to assay setup. For ADCC specificity assessment, NK cell effectors remained unstained, with targets composed of a Miltenyi CD34 magnetic bead–selected mixture of Q8- and RQR8-transduced T cells in equal proportion with Q8 and RQR8 targets identified by expression of eBFP2 or eGFP fluorescent proteins, respectively.
Chromium release cytotoxicity assay
Cytotoxicity of CAR T cells was evaluated in a standard 4-hour 51Cr release assay as previously described.22
In vivo engraftment model
This work was performed under United Kingdom home-office–approved project license and in accordance with institutional policies. Splenocytes from C57BL/6 mice were transduced with SFGmR.RQR8 in the absence of stimulation following 24 hours of stimulation by Con A/IL7 (2 µg/mL and 1 ng/mL, respectively). Following transduction, splenocytes were restimulated with IL-2 (50 ng/mL) and cultured overnight. Following a single round of Miltenyi CD34 selection, purity >90% was achieved. A total of 1.5 × 106 transgenic splenocytes were administered by tail-vein injection to (C57BL/6 × BALB/c) F1 recipients 4 hours following 5-Gy radiograph irradiation preconditioning. Engraftment was assessed 7 days posttransfer by QBEnd10 staining of peripheral blood. A total of 150 µg of murine IgG2a isotyped rituximab was administered by tail-vein injection on days 7, 10, and 12 posttransfer. Mice were sacrificed on day 14 with samples taken from peripheral blood, spleen, lymph nodes, and bone marrow stained for CD4, CD8, H2Kd, and QBEnd10 to assess engraftment.
Results
The monoclonal antibody QBEnd10 binds a linear 16-residue fragment of CD34
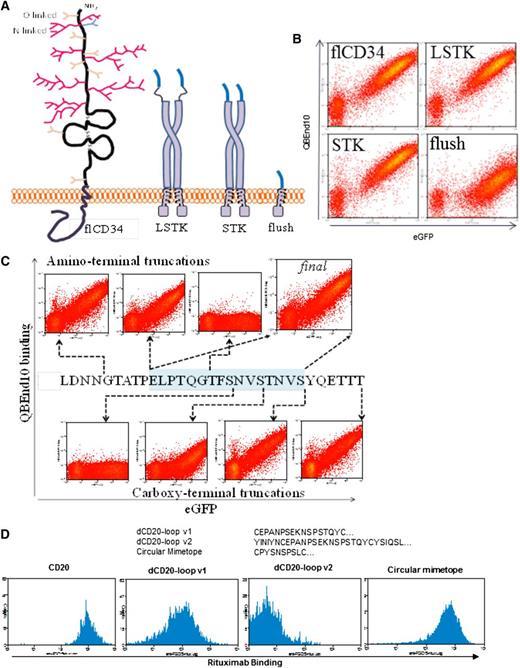
We first identified the binding site of the type I anti-CD34 monoclonal antibody (mAb) QBEnd10. A DNA library coding for overlapping 31-residue fragments of the serine/threonine-rich amino terminus of CD34 was generated. This library was cloned into a retroviral vector between CD34 signal peptide and transmembrane domain. SupT1 cells were transduced with the subsequent retroviral library. QBEnd10 binding cells were single-cell sorted and the retroviral integrant subsequently sequenced from genomic DNA (not shown). From this, we determined that QBEnd10 binds to the amino-terminal 40 amino acids of mature CD34. We hypothesized that this epitope might need to be projected from the cell surface and allowed loose orientation to allow effective QBEnd10 binding. The CD8 stalk is composed of only 42 amino acids, but it results in a projection sufficient to allow the globular CD8 amino terminus to reach past the TCR and interact with major histocompatibility complex class I on a neighboring cell.23 By transducing SupT1 cells with equal titer of retroviral supernatant, we then compared QBEnd10 binding to full-length CD34 with the above 40-residue epitope connected directly to the CD8 transmembrane domain, fused directly to the CD8 stalk, or connected to the CD8 stalk via a serine-glycine linker. Both constructs with the CD8 stalk exhibited similar binding to full-length CD34, suggesting that the CD8 stalk enhances access of QBEnd10 to the epitope (Figure 1A-B). Finally, we refined the epitope further by sequential deletion of the amino and carboxy termini. We concluded that a 16-residue linear sequence of CD34 on a CD8 stalk could recapitulate equivalent binding of QBEnd10 as the full CD34 protein (Figure 1C).
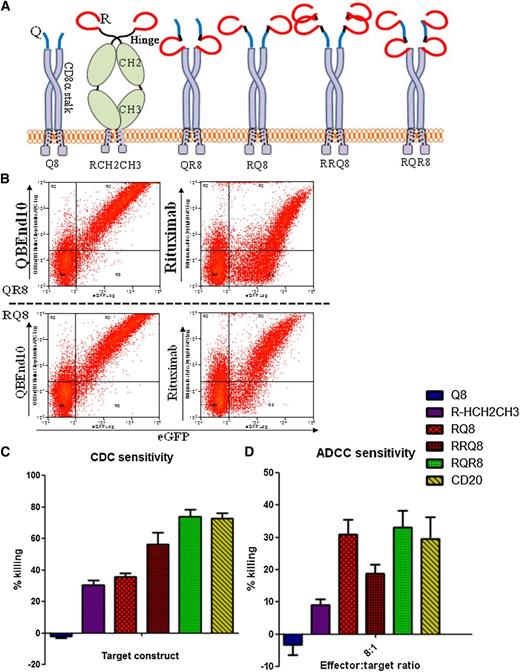
Engineering of QBEnd10- and rituximab-binding domains. (A) Coding sequences for the 31 extreme amino-terminal residues of mature full-length CD34 (flCD3) were cloned in-frame to the CD8 stalk and transmembrane domain via a serine-glycine linker (LSTK) or without this linker (STK) or connected directly to the CD8 transmembrane domain (flush). These open-reading frames were coexpressed with eGFP within a bicistronic retroviral vector. (B) Flow cytometric analysis of eGFP and QBEnd10 staining in T cells transduced with flCD34, LSTK, STK. QBEnd10 binding equivalent to that of flCD34 was seen in constructs containing the CD8 stalk, but not with the flush construct. The serine glycine linker did not improve QBEnd10 binding. (C) Further epitope minimization was performed by sequential amino- and carboxy-terminal deletion of the 31 residues of CD34 until binding of QBEnd10 was abrogated. In this way, we established a final minimal epitope-binding construct containing only 16-amino-acid residues from the 385 present in the native antigen. (D) Binding of rituximab to T cells transduced with full-length CD20 (CD20), the major extracellular loop of CD20 with some flanking residues on the CD8 stalk (dCD20-loop v2), the major extracellular loop of CD20 delineated precisely at the constraining cysteine on a CD8 stalk (dCD20-loop v1), and the circular CD20 mimotope described by Perosa et al26 on a CD8 stalk.
Engineering of QBEnd10- and rituximab-binding domains. (A) Coding sequences for the 31 extreme amino-terminal residues of mature full-length CD34 (flCD3) were cloned in-frame to the CD8 stalk and transmembrane domain via a serine-glycine linker (LSTK) or without this linker (STK) or connected directly to the CD8 transmembrane domain (flush). These open-reading frames were coexpressed with eGFP within a bicistronic retroviral vector. (B) Flow cytometric analysis of eGFP and QBEnd10 staining in T cells transduced with flCD34, LSTK, STK. QBEnd10 binding equivalent to that of flCD34 was seen in constructs containing the CD8 stalk, but not with the flush construct. The serine glycine linker did not improve QBEnd10 binding. (C) Further epitope minimization was performed by sequential amino- and carboxy-terminal deletion of the 31 residues of CD34 until binding of QBEnd10 was abrogated. In this way, we established a final minimal epitope-binding construct containing only 16-amino-acid residues from the 385 present in the native antigen. (D) Binding of rituximab to T cells transduced with full-length CD20 (CD20), the major extracellular loop of CD20 with some flanking residues on the CD8 stalk (dCD20-loop v2), the major extracellular loop of CD20 delineated precisely at the constraining cysteine on a CD8 stalk (dCD20-loop v1), and the circular CD20 mimotope described by Perosa et al26 on a CD8 stalk.
Rituximab binding equivalent to that of full-length CD20 could only be achieved with a mimotope
We next sought to achieve binding by rituximab to a fragment of CD20 in an analogous fashion. The binding site of rituximab to CD20 has been elucidated by deletion studies24 and crystallography25 ; rituximab binds to the disulfide-constrained portion of the CD20 major extracellular loop. We generated constructs where this disulfide-constrained loop was expressed on the CD8 stalk with no flanking residues (dCD20-loop v1) or 4 additional residues of CD20 on either side (dCD20-loop v2). Construct dCD20-loop v2 did not bind rituximab, whereas dCD20-loop v1 achieved only very poor binding (Figure 1D). Perosa et al26 described rituximab-binding mimotopes of CD20. These sequences resemble the major extracellular loop of CD20 and compete with rituximab for CD20 binding. Hence, we next generated constructs connecting these mimotopes to the CD8 stalk, connecting a circular mimotope to the CD8 stalk. This could recapitulate binding of rituximab. We next generated constructs with the QBEnd10 epitope linked to the CD20 mimotope in either orientation (constructs termed RQ8 and QR8, respectively) (Figure 2A). This circular mimotope and the QBEnd10 epitope described above could be coexpressed in either orientation on CD8 stalk with no diminution of binding of either antibody (Figure 2B).
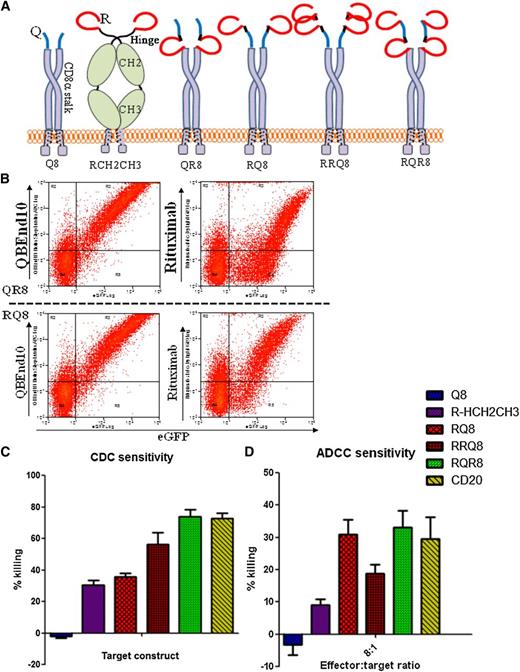
Engineering of combination QBEnd10/rituximab-binding constructs. (A) Cartoon of alternative epitope constructs illustrating alternative presentation formats of epitope constructs that were generated to enable comparison of sensitivity to CDC- and ADCC-mediated deletion. “Q” refers to the 16-amino-acid minimized QBEnd10 epitope; “R” refers to the CD20 circular mimotope; “8” refers to the CD8α stalk; and “Hinge-CH2CH3” refers to the hinge, CH2 and CH3, domains of human IgG1. (B) Binding of QBEnd10 and rituximab to constructs QR8 and RQ8 coexpressed with eGFP in primary human T cells is shown. Both antibodies could bind in either orientation. (C) Sensitivity to CDC depletion using primary human T cells transduced with constructs Q8, RCH2CH3, RQ8, RRQ8, RQR8, and full-length CD20 is shown. Following 4-hour incubation with 25% baby-rabbit complement and rituximab at 100 μg/mL, samples were stained with Annexin V/PI and the live population was assessed by flow cytometry analysis for the presence of the coexpressed eGFP marker gene. Results illustrate comparative deletion observed from 3 separate donors. (D) Similarly, sensitivity to ADCC-mediated depletion was assessed using primary human T-cell targets transduced with constructs were challenged by 16:1, 8:1, 4:1, and 2:1 effector:target ratios of NK cell effectors derived from the same donor. Following 48-hour incubation in the presence of 100 μg/mL rituximab, depletion was assessed by flow cytometry analysis. Samples were stained with Annexin V/PI for live/dead exclusion, with NK cells labeled with CellTRACE violet excluded from the live gate to identify the residual live population of targets cells identified by the presence of a coexpressed eGFP marker gene. Results illustrate comparative deletion by the 8:1 effector:target ratio observed from 3 separate donors. Construct RQR8 engenders equal CDC and ADCC to full-length CD20.
Engineering of combination QBEnd10/rituximab-binding constructs. (A) Cartoon of alternative epitope constructs illustrating alternative presentation formats of epitope constructs that were generated to enable comparison of sensitivity to CDC- and ADCC-mediated deletion. “Q” refers to the 16-amino-acid minimized QBEnd10 epitope; “R” refers to the CD20 circular mimotope; “8” refers to the CD8α stalk; and “Hinge-CH2CH3” refers to the hinge, CH2 and CH3, domains of human IgG1. (B) Binding of QBEnd10 and rituximab to constructs QR8 and RQ8 coexpressed with eGFP in primary human T cells is shown. Both antibodies could bind in either orientation. (C) Sensitivity to CDC depletion using primary human T cells transduced with constructs Q8, RCH2CH3, RQ8, RRQ8, RQR8, and full-length CD20 is shown. Following 4-hour incubation with 25% baby-rabbit complement and rituximab at 100 μg/mL, samples were stained with Annexin V/PI and the live population was assessed by flow cytometry analysis for the presence of the coexpressed eGFP marker gene. Results illustrate comparative deletion observed from 3 separate donors. (D) Similarly, sensitivity to ADCC-mediated depletion was assessed using primary human T-cell targets transduced with constructs were challenged by 16:1, 8:1, 4:1, and 2:1 effector:target ratios of NK cell effectors derived from the same donor. Following 48-hour incubation in the presence of 100 μg/mL rituximab, depletion was assessed by flow cytometry analysis. Samples were stained with Annexin V/PI for live/dead exclusion, with NK cells labeled with CellTRACE violet excluded from the live gate to identify the residual live population of targets cells identified by the presence of a coexpressed eGFP marker gene. Results illustrate comparative deletion by the 8:1 effector:target ratio observed from 3 separate donors. Construct RQR8 engenders equal CDC and ADCC to full-length CD20.
Two copies of rituximab mimotope flanking the QBEnd10 epitope on the CD8 are optimal for rituximab-mediated lysis
Despite binding of rituximab, CDC assays with RQ8 and QR8 showed significantly less lysis than with full-length CD20. We next proceeded to try to gain some insight into why this was and to improve killing. The circular rituximab mimotope was placed into several formats. RCH2CH3 was constructed by cloning the mimotope onto a human IgG1 Fc spacer. RRQ8 was constructed by linking 2 copies of the rituximab mimotope followed by the QBEnd10 epitope and CD8 stalk. RQR8 has 2 copies of the rituximab mimotope flanking the QBEnd10 epitope on the CD8 stalk. (Figure 2A). Primary human T cells were transduced with these constructs, along with Q8 and full-length codon-optimized CD20. CDC and ADCC assays were performed on unsorted T-cell populations. Constructs RQ8 and RQR8 resulted in ADCC equal to that of CD20, whereas only RQR8 resulted in CDC equal to that of the full-length protein (P < .05). We hence selected RQR8 for further study.
RQR8 has properties that make it a good marker/suicide gene
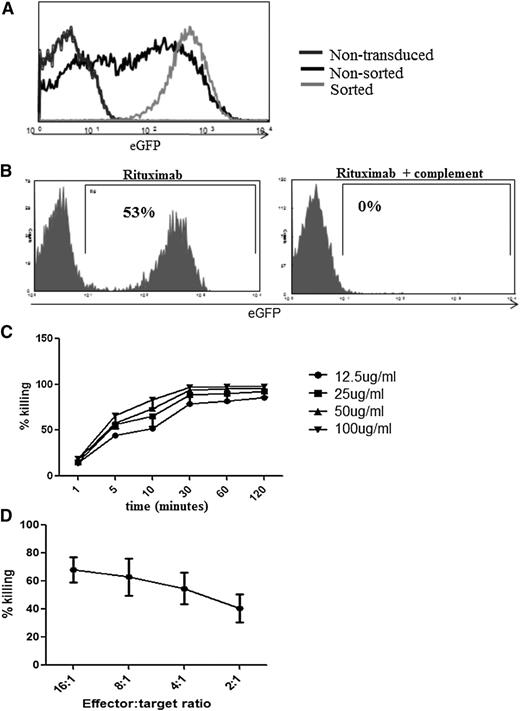
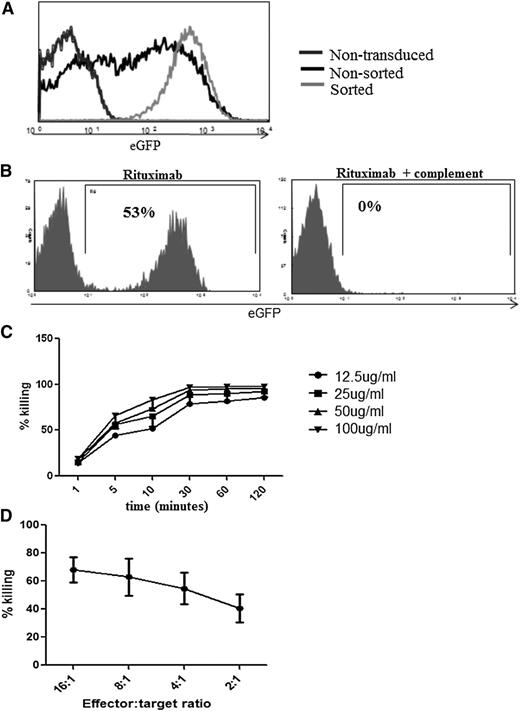
We first explored utility of RQR8 as a marker gene. Primary human T cells transduced with RQR8 could be sorted to high stringency with Miltenyi CD34 beads (Figure 3A). Further, as few as 0.01% RQR8-transduced primary human T cells could be detected in serial dilution experiments (supplemental Figure 1). Further, RQR8 allows immunohistochemical detection with QBEnd10. Concomitant staining for CD3 or with another CD34 antibody will easily discriminate transgenic T cells from other CD34 cells such as hematopoietic stem cells or endothelium (supplementary Figure 2). Next, we explored the utility of RQR8 as a suicide gene. Miltenyi QBEnd10 bead-sorted T cells are very efficiently lysed by a single exposure to rituximab and complement. In 50:50 mixing assays, sorted T-cells were mixed back with nontransduced T cells, and 98% of transduced T cells could be depleted by rituximab and complement (Figure 3B). Both CDC- and ADCC-mediated sensitivities of RQR8 are equal to that demonstrated by expression of full-length codon-optimized CD20 (Figure 2B-C). Rituximab-mediated deletion is efficient at 25 µg/mL, which is well within the therapeutic window of this agent (Figure 3C).27 CDC-mediated killing of RQR8-expressing T cells occurs rapidly, with maximum lysis within 30 minutes of exposure (Figure 3C).
Functional characterization of RQR8. (A) Demonstration of typical cellular purification result achieved following CD34 magnetic bead selection. Primary human T cells were transduced with the bicistronic retroviral vector SFG.RQR8.IRES.eGFP and selected with Miltenyi QBEnd10 beads. (B) Efficiency of CDC in transduced sorted T cells. Primary human T cells were transduced with SFG.RQR8.IRES.eGFP, purified with QBEnd10 beads, and combined at equal concentration with nontransduced T cells. This mixed population was exposed to a 2-hour incubation with 100 μg/mL rituximab with or without 25% baby-rabbit complement. T cells were stained with Annexin/PI, and flow cytometric analysis of eGFP expression on the live population is shown. This is an example of an experiment repeated 6 times in different donors. More than 95% of the transduced population is deleted. (C) Time course and rituximab dose-titration assay. CDC-mediated deletion of targets was performed in primary human T cells with rituximab concentrations of 12.5, 25, 50, and 100 μg/mL analyzed at 1, 5, 10, 30, 60, and 120 minutes. Figure shows mean and standard deviation from 3 donors. CDC is highly effective at rituximab concentrations of ≥25 μg/mL, and killing occurs within 30 minutes. (D) Demonstration of ADCC-mediated sensitivity against T cells transduced with RQR8. Transduced T cells were incubated at 16:1, 8:1, 4:1, and 2:1 effector:target ratios of NK cell effectors derived from the same donor exposed to 100 μg/mL rituximab. Samples were stained with Annexin V/PI for live/dead exclusion with depletion assessed by flow cytometry analysis comparison of the ratio of eGFP/eBFP2 marker gene expression from the residual live population. Note: QR8 was not assessed for ADCC sensitivity.
Functional characterization of RQR8. (A) Demonstration of typical cellular purification result achieved following CD34 magnetic bead selection. Primary human T cells were transduced with the bicistronic retroviral vector SFG.RQR8.IRES.eGFP and selected with Miltenyi QBEnd10 beads. (B) Efficiency of CDC in transduced sorted T cells. Primary human T cells were transduced with SFG.RQR8.IRES.eGFP, purified with QBEnd10 beads, and combined at equal concentration with nontransduced T cells. This mixed population was exposed to a 2-hour incubation with 100 μg/mL rituximab with or without 25% baby-rabbit complement. T cells were stained with Annexin/PI, and flow cytometric analysis of eGFP expression on the live population is shown. This is an example of an experiment repeated 6 times in different donors. More than 95% of the transduced population is deleted. (C) Time course and rituximab dose-titration assay. CDC-mediated deletion of targets was performed in primary human T cells with rituximab concentrations of 12.5, 25, 50, and 100 μg/mL analyzed at 1, 5, 10, 30, 60, and 120 minutes. Figure shows mean and standard deviation from 3 donors. CDC is highly effective at rituximab concentrations of ≥25 μg/mL, and killing occurs within 30 minutes. (D) Demonstration of ADCC-mediated sensitivity against T cells transduced with RQR8. Transduced T cells were incubated at 16:1, 8:1, 4:1, and 2:1 effector:target ratios of NK cell effectors derived from the same donor exposed to 100 μg/mL rituximab. Samples were stained with Annexin V/PI for live/dead exclusion with depletion assessed by flow cytometry analysis comparison of the ratio of eGFP/eBFP2 marker gene expression from the residual live population. Note: QR8 was not assessed for ADCC sensitivity.
RQR8 is of utility when expressed with CARs
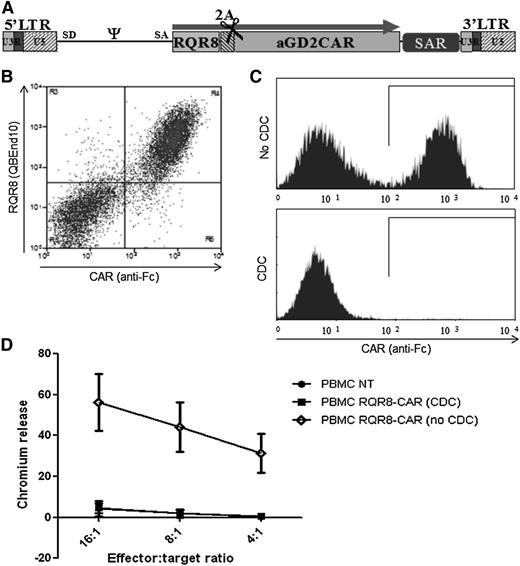
CARs are among the most commonly used T-cell engineering components. We coexpressed RQR8 with an anti-GD2 CAR in a retroviral vector using the self-cleaving FMD-2A sequence.17 This resulted in obligate coexpression of CAR and RQR8 (Figure 4B). Selection with QBEnd10 beads resulted in a pure population of CAR-expressing T cells (>95% from 6 separate experiments). We next mixed RQR8-sorted T cells 1:1 with nontransduced T cells and performed lysis and functional experiments. CDC by rituximab led to deletion of >97% of the CAR-expressing population (Figure 4C). Further, this depleted population completely lost recognition of antigen-expressing targets (Figure 4D). RQR8 could also be coexpressed in a tricistronic vector with a native α/β TCR with FMD-2A–like sequences (not shown).
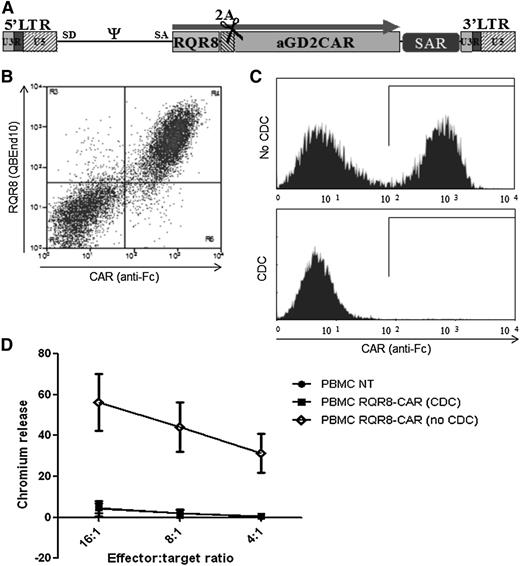
Demonstration of RQR8 activity when coexpressed with a CAR. (A) RQR8 is expressed with an anti-GD2 CAR in the SFG retroviral vector using the FMD-2A sequence. The retroviral vector expression is enhanced using a scaffold attachment region (SAR). (B) Primary human T cells were transduced with this retroviral vector. Following transduction, cells were stained with QBEnd10 and polyclonal goat anti-human Fc (the latter stains the CAR). Clear coexpression of RQR8 and CAR was observed. (C) T cells were sorted with QBEnd10 beads and mixed 1:1 with nontransduced T cells and exposed to either rituximab alone or rituximab and complement. Two hours later, anti-Fc and Annexin V staining was performed. Anti-Fc staining is shown gating on live T cells. Mean depletion was 97% of transduced cells. (D) This rituximab-depleted population, along with the undepleted T cells, and nontransduced T-cells were used as effectors in a chromium-release killing assay using GD2-positive target cells. This experiment was performed in 5 donors. LTR, long terminal repeat.
Demonstration of RQR8 activity when coexpressed with a CAR. (A) RQR8 is expressed with an anti-GD2 CAR in the SFG retroviral vector using the FMD-2A sequence. The retroviral vector expression is enhanced using a scaffold attachment region (SAR). (B) Primary human T cells were transduced with this retroviral vector. Following transduction, cells were stained with QBEnd10 and polyclonal goat anti-human Fc (the latter stains the CAR). Clear coexpression of RQR8 and CAR was observed. (C) T cells were sorted with QBEnd10 beads and mixed 1:1 with nontransduced T cells and exposed to either rituximab alone or rituximab and complement. Two hours later, anti-Fc and Annexin V staining was performed. Anti-Fc staining is shown gating on live T cells. Mean depletion was 97% of transduced cells. (D) This rituximab-depleted population, along with the undepleted T cells, and nontransduced T-cells were used as effectors in a chromium-release killing assay using GD2-positive target cells. This experiment was performed in 5 donors. LTR, long terminal repeat.
T cells expressing RQR8 are not susceptible to lysis by ofatumumab
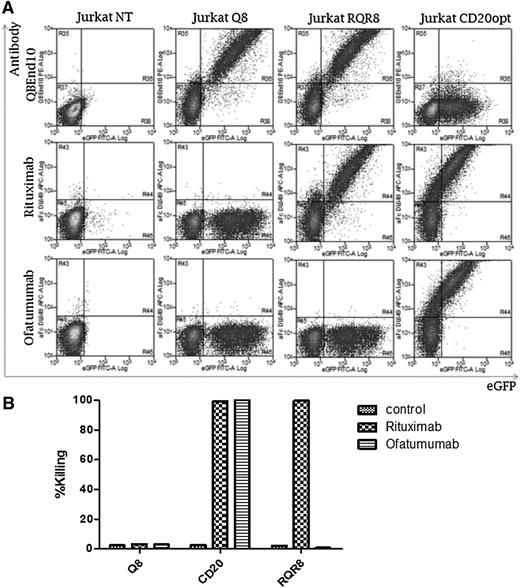
Recent clinical studies of adoptive immunotherapy with CAR-engineered T cells have focused on B-cell malignancies. In this setting, concomitant administration of a B-cell–depleting therapeutic antibody might be desirable. Given the long in vivo half-life of rituximab, its inclusion in a conditioning regimen precludes use of RQR8 in the vector. However, a new generation of anti-CD20 therapeutic antibodies has been developed. One such agent, ofatumumab, binds to a different epitope on CD20 than that of rituximab.28 We investigated whether ofatumumab would bind to and cause lysis of RQR8-expressing cells. First, T cells transduced with Q8, CD20, and RQR8 were stained with rituximab and ofatumumab (Figure 5A). Whereas CD20-expressing T cells bound both antibodies, RQR8-expressing T cells only bound rituximab. Further, a CDC assay showed lysis of CD20-expressing T cells with both ofatumumab and rituximab (Figure 5B), whereas only rituximab was capable of lysing RQR8-expressing T cells. Hence, ofatumumab can be used in a preparative regimen of engineered T-cell therapy with RQR8. Binding of a panel of other anti-CD20 mAbs in clinical use or preclinical development to RQR8 is detailed in supplemental Table 1.
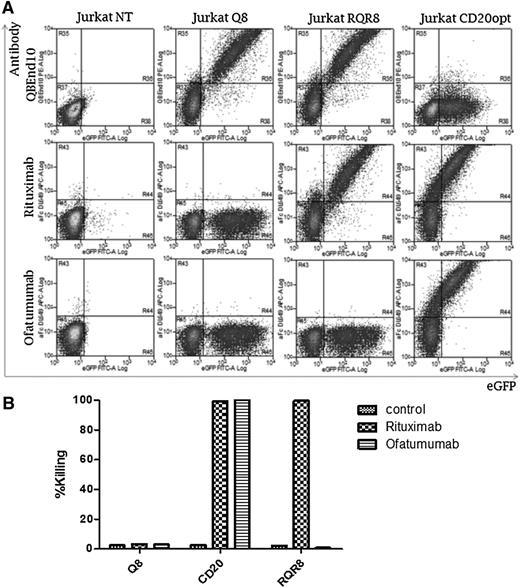
Binding and killing with ofatumumab vs rituximab. T cells well transduced to express Q8, RQR8, or codon-optimized CD20, all coexpressed with eGFP. (A) T cells were stained with either rituximab or ofatumumab and a secondary anti-human-Fc allophycocyanin-conjugated goat antibody. Antibody binding vs eGFP expression is shown. Q8 binds neither mAb; CD20 binds both, whereas RQR8 only binds rituximab. (B) Next, primary human T cells were transduced with these constructs and a CDC assay was performed with either rituximab or ofatumumab. The percentage lysis as determined by Annexin-V and PI positivity is shown. Q8 transduced to T cells was not lysed by either mAb; T cells expressing CD20 were lysed by both, whereas RQR8 T cells were lysed only by rituximab. This experiment was performed in 1 donor.
Binding and killing with ofatumumab vs rituximab. T cells well transduced to express Q8, RQR8, or codon-optimized CD20, all coexpressed with eGFP. (A) T cells were stained with either rituximab or ofatumumab and a secondary anti-human-Fc allophycocyanin-conjugated goat antibody. Antibody binding vs eGFP expression is shown. Q8 binds neither mAb; CD20 binds both, whereas RQR8 only binds rituximab. (B) Next, primary human T cells were transduced with these constructs and a CDC assay was performed with either rituximab or ofatumumab. The percentage lysis as determined by Annexin-V and PI positivity is shown. Q8 transduced to T cells was not lysed by either mAb; T cells expressing CD20 were lysed by both, whereas RQR8 T cells were lysed only by rituximab. This experiment was performed in 1 donor.
T cells transduced with RQR8 can be efficiently deleted in vivo
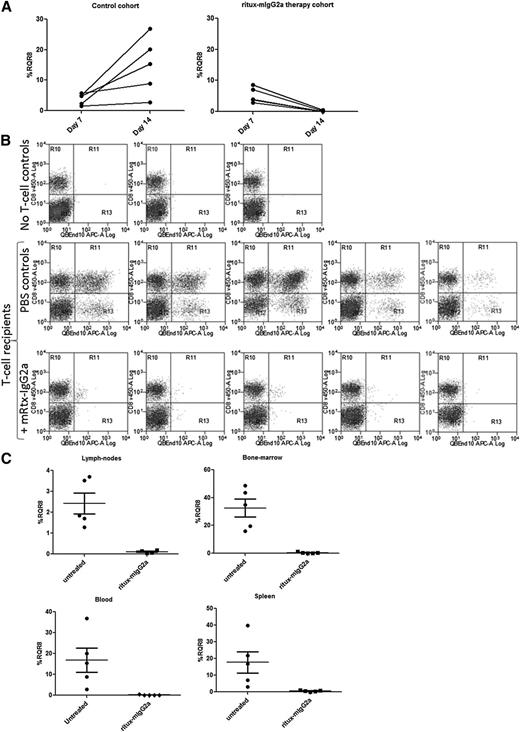
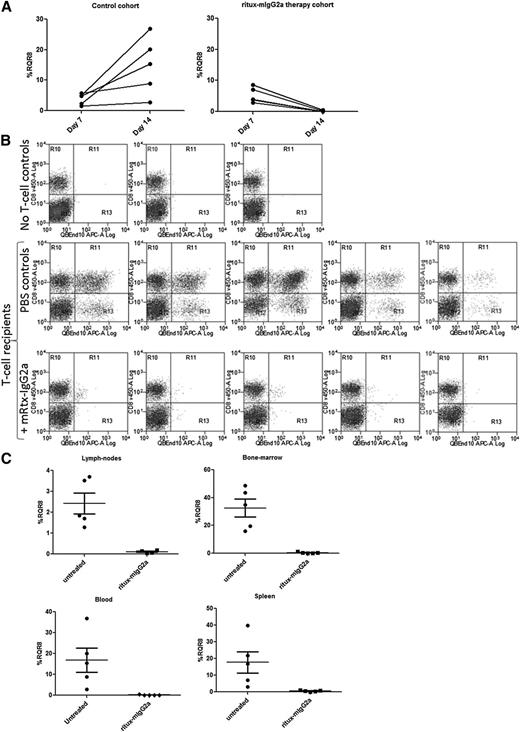
We used an immunocompetent haploidentical adoptive transfer model with RQR8-transduced C57BL/6 splenocytes transferred to non–lethally irradiated C57BL/6 × BALB/c cross (F1) recipients. This model allows good engraftment levels in all lymphoid tissue sustained by allogeneic stimulation, but it also preserves endogenous lymphocytes. Rituximab is a chimeric antibody with mouse variable regions and human κ/IgG1 constant regions with limited activity in mice. To more truly determine potency of deletion, we reengineered rituximab to mouse IgG2a, the functional equivalent of human IgG129 (referred henceforth as ritux-mIgG2a; see supplemental Data 3 “Materials and methods” and supplemental Figure 3). A total of 1.5 × 106 RQR8-transduced QBEnd10-sorted splenocytes resulted in engraftment readily detectable at day 7 in all mice. After 1 week of ritux-mIgG2a administration, transgenic T cells were no longer detectable in peripheral blood by flow cytometry (P < .001), whereas marking levels continued to increase in the control cohort (Figure 6A). Mice were then sacrificed and assessed for engraftment in lymph nodes, bone marrow, and spleen. Depletion levels of 96%, 98%, and 99% were observed (P < .001) in blood, bone marrow, lymph nodes, and spleen (Figure 6B-C). This depletion was in face of the considerable engraftment evident in bone marrow (mean 35%) and spleen (mean 22%) (Figure 6C). RQR8 allows for rapid and highly effective in vivo depletion of transgenic T cells in all lymphoid compartments. In addition, we performed an in vivo time course with separate cohorts of mice sacrificed at different time points. Supplemental Figure 4 shows depletion at 6, 48, and 168 hours in peripheral blood within individual mice. Supplemental Figure 5 shows depletion in blood, spleen, bone marrow, and lymph nodes in different cohorts of mice at 6, 24, 48, 120, and 168 hour time points. This demonstrates over 50%, 60%, and 70% depletion in spleen, bone marrow, and blood, respectively, within 6 hours after rituximab therapy. Dynamics of increasing engraftment in control animals and increasingly effective depletion in ritux-IgG2a recipients is observed.
In vivo testing of RQR8. C57BL/6 splenocytes were retrovirally transduced to express RQR8 and selected to >90% purity with Miltenyi CD34 beads. Approximately 1.5 million of these cells were injected IV into 5-Gy radiograph preconditioned C57BL/6 × BALB/c cross (F1) recipients. Seven days posttransfer, engraftment was assessed by peripheral blood. ritux-mIgG2a therapy commenced with 150-μg doses at days 7, 10, and 12 by IV injection or phosphate-buffered saline carrier for the control cohort. Each cohort had 5 mice. An additional 2 mice were irradiated but given neither T cells nor ritux-mIgG2a. Animals were sacrificed at day 14, and engraftment was assessed by considering the proportion of QBEnd10-positive T cells as a proportion of the T-cell population. (A) Peripheral blood marking levels in control and in treated mice. (B) Flow cytometry of the T-cell population in spleen showing RQR8 vs CD8 in mice not receiving transgenic T cells and control and rituximab-treated mice. (C) Percentage marking in lymph nodes, bone marrow, blood, and spleen.
In vivo testing of RQR8. C57BL/6 splenocytes were retrovirally transduced to express RQR8 and selected to >90% purity with Miltenyi CD34 beads. Approximately 1.5 million of these cells were injected IV into 5-Gy radiograph preconditioned C57BL/6 × BALB/c cross (F1) recipients. Seven days posttransfer, engraftment was assessed by peripheral blood. ritux-mIgG2a therapy commenced with 150-μg doses at days 7, 10, and 12 by IV injection or phosphate-buffered saline carrier for the control cohort. Each cohort had 5 mice. An additional 2 mice were irradiated but given neither T cells nor ritux-mIgG2a. Animals were sacrificed at day 14, and engraftment was assessed by considering the proportion of QBEnd10-positive T cells as a proportion of the T-cell population. (A) Peripheral blood marking levels in control and in treated mice. (B) Flow cytometry of the T-cell population in spleen showing RQR8 vs CD8 in mice not receiving transgenic T cells and control and rituximab-treated mice. (C) Percentage marking in lymph nodes, bone marrow, blood, and spleen.
Discussion
Genetic engineering of T cells is increasing the scope and application of adoptive immunotherapy. Unlike traditional small-molecule or protein therapeutics, adoptive immunotherapy aims to achieve engraftment of autonomous cells with the capacity to persist indefinitely and proliferate. Hence, toxicity may be long-lived and may escalate. Further, cellular therapies are highly complex, and optimization of several parameters such as transgene and cellular production with rapid iterative clinical experiments are likely needed before development of phase 2/3 studies. A simple compact T-cell engineering component that facilitates production and allows selective depletion in clinical study subjects without requirement for new clinical-grade reagents would be of value to the field.
Existing strategies that have been clinically tested to allow selective depletion of transgenic T cells (suicide genes) include the HSV-TK gene and iCasp9. The former renders T cells susceptible to ganciclovir, whereas the latter22 is activated by a small-molecular chemical inducer of dimerization. HSV-TK is an effective suicide gene,10 but preformed immune responses may restrict its use to clinical settings where there is marked immunosuppression, such as in haploidentical stem cell transplant recipients30,31 iCasp9 is also effective, but its use is dependent on the availability of a proprietary chemical inducer of dimerization manufactured to clinical grade.
Both HSV-TK and iCasp9 are intracellular proteins, which prevents their use as simultaneous marker genes that would allow tracking engraftment and the selection and purification of transduced cells prior to administration. Truncated CD34 and truncated CD19 have been coexpressed with HSV-TK and iCasp9, respectively, as marker genes, and both allow clinical-grade Miltenyi CliniMACS sorting of transduced T cells as well as tracking of transduced cells by flow cytometry in samples from study subjects. However, CD19 is currently the prime target for CAR therapy, and using this antigen as a marker precludes concomitant targeting. Truncated or full-length CD34 may impact on the migratory ability of modified cells.32 Further, a combined marker and suicide gene adds a considerable genetic payload to the gene vector.
Therefore, we reasoned that we could construct a highly compact combined sort-suicide gene using antibody-binding epitopes from both CD34 and CD20. Further, by aiming to recapitulate the properties of CD34 as a marker gene and CD20 as a suicide gene, our construct could be used with “off-the-shelf” reagents. First, we determined that the epitope of CD34 recognized by QBEnd10 was a linear peptide localized to the extreme amino terminus of the protein. The crystallographic data of the CD20 molecule suggested that the major extracellular loop was all that was required for binding to rituximab, and we were surprised when this loop failed to afford full binding. We concluded that correct orientation of this loop only occurs in full-length CD20, and replacing this loop with a mimotope afforded a solution. The need for 2 copies of the rituximab-binding mimotope to result in effective CDC may be explained by the requirement to cluster CD20 to effect lysis. Our final homodimeric (and hence tetravalent) RQR8 allows crosslinking and clustering that may resemble that seen by the native antigen, CD20.
With all nonnative genes used to engineer T cells, immunogenicity presents a potential limitation. For instance, CARs are typically derived from murine antibodies and have many junctional and linker sequences exposed. Similarly, RQR8 has junctional sequences that may be immunogenic. A bioinformatics analysis of B- and T-cell immunogenicity of RQR8 is presented in supplemental Figure 6, identifying 6 T-cell and 24 B-cell epitopes. It should be noted that current CAR and TCR adoptive immunotherapy protocols typically employ profoundly lymphodepletion preparatory regimens resulting in profound immunosuppression at the time of T-cell administration. This may reduce the occurrence of, and may prevent development of, an immune response. Ultimately, only clinical studies can determine the practical consequences of immunogenicity of any particular T-cell engineering component.
Suicide-gene strategies based on transgenic expression of a binding target for a therapeutic antibody have been proposed,33-36 with a cetuximab-based system34 being the closest to RQR8. Here, truncated epidermal growth factor receptor (EGFR) is expressed on a T cell and depletion effected by the anti-EGFR therapeutic mAb cetuximab. This system is less likely to be immunogenic than RQR8. However, RQR8 has some advantages. Firstly, RQR8 has an off-the-shelf clinical grade selection system available. Secondly, rituximab monotherapy is well tolerated by the majority of patients with little increase in opportunistic infection37,38 and no maximally tolerated dose. In contrast, treatment with cetuximab is accompanied by acneiform follicular skin exanthema in >80% of patients. Severe exanthema (grade III/IV) develops in about 9% to 19% of patients with the necessity of cetuximab dose reduction or cessation.39 Thirdly, rituximab’s property as a highly potent lymphodepleting agent in human subjects is well established, whereas given its target, no such data exist with cetuximab. Finally, RQR8 is 136 amino acids vs truncated EGFR’s 336 residues.
In summary, we have created a 136-amino-acid epitope-based marker/suicide gene for T cells. The translated protein is stably expressed on the cell surface following retroviral transduction. It binds QBEnd10 analogously to full-length CD34, allowing clinical-grade sorting with off-the-shelf reagents and easy in vivo tracking. Further, the construct binds rituximab; the dual-epitope design engenders highly effective CDC and ADCC and consequently renders T cells highly susceptible to in vivo rituximab-mediated depletion. Due to the small size of RQR8, it can easily be coexpressed with a wide range of T-cell engineering components. We anticipate this construct will be of great practical utility and render T-cell therapy safer and ultimately more efficacious.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mark Craggs and Martin Glennie (University of Southampton) for assistance and helpful discussions and Arnold Pizzey for assistance with flow cytometry.
This work was supported by a Medical Research Council Clinician Scientist Fellowship (M.P.), the European Union Framework 6 Consortium “CHILDHOPE”, a Leukaemia and Lymphoma Research project grant, and the United Kingdom National Institute for Health Research University College London Hospital Biomedical Research Centre.
Authorship
Contribution: M.P. conceived the idea and designed initial constructs, designed subsequent experiments, and wrote the manuscript; B.P. designed the final construct, codesigned most experiments, performed most of the molecular cloning, retroviral, and in vitro and in vivo work, and assisted in writing the manuscript; L.M. assisted with in vivo work; S.T. assisted with molecular cloning; E.K. assisted with molecular cloning and in vitro and in vivo work; K.S. assisted with in vitro work; D.L. assisted in the design of experiments and writing the manuscript; T.M. performed the immunohistochemistry; R.C., B.F., K.P., and S.Q. advised and assisted with animal work; and V.M. assisted with some of the molecular cloning.
Conflict-of-interest disclosure: D.L. is on the scientific advisory board of Cellectis Therapeutics, which is licensing RQR8. The laboratories of M.P. and K.P. receive funding for contract research from Cellectis Therapeutics. The remaining authors declare no competing interests.
Correspondence: Martin Pule, UCL Cancer Institute, University College London, Paul O’Gorman Building, 72 Huntley St, London WC1E 6HX, United Kingdom; e-mail: m.pule@ucl.ac.uk.