Key Points
HLA-DRB1*07:01 is associated with asparaginase hypersensitivity and anti-asparaginase antibodies.
HLA-DRB1 alleles that confer high-affinity binding to asparaginase epitopes lead to a higher frequency of hypersensitivity.
Abstract
Asparaginase is a therapeutic enzyme used to treat leukemia and lymphoma, with immune responses resulting in suboptimal drug exposure and a greater risk of relapse. To elucidate whether there is a genetic component to the mechanism of asparaginase-induced immune responses, we imputed human leukocyte antigen (HLA) alleles in patients of European ancestry enrolled on leukemia trials at St. Jude Children’s Research Hospital (n = 541) and the Children’s Oncology Group (n = 1329). We identified a higher incidence of hypersensitivity and anti-asparaginase antibodies in patients with HLA-DRB1*07:01 alleles (P = 7.5 × 10−5, odds ratio [OR] = 1.64; P = 1.4 × 10−5, OR = 2.92, respectively). Structural analysis revealed that high-risk amino acids were located within the binding pocket of the HLA protein, possibly affecting the interaction between asparaginase epitopes and the HLA-DRB1 protein. Using a sequence-based consensus approach, we predicted the binding affinity of HLA-DRB1 alleles for asparaginase epitopes, and patients whose HLA genetics predicted high-affinity binding had more allergy (P = 3.3 × 10−4, OR = 1.38). Our results suggest a mechanism of allergy whereby HLA-DRB1 alleles that confer high-affinity binding to asparaginase epitopes lead to a higher frequency of reactions. These trials were registered at www.clinicaltrials.gov as NCT00137111, NCT00549848, NCT00005603, and NCT00075725.
Introduction
Asparaginase is used to treat leukemias and lymphomas and is one of the essential anticancer drugs for the curative treatment of acute lymphoblastic leukemia (ALL). The drug, a nonhuman enzyme, depletes asparagine in serum and inhibits the proliferation of leukemic cells; however, the precise mechanism of asparaginase action is not completely understood.1 Immune responses to asparaginase during treatment are common. About 30% to 75% of patients receiving native Escherichia coli asparaginase experience hypersensitivity reactions manifesting typically as urticaria and wheezing, and as many as 70% develop anti-asparaginase antibodies after administration.2-5 Furthermore, patients with lower exposure to asparaginase because of hypersensitivity reactions or the development of neutralizing antibodies, have worse outcomes compared with patients with fewer reactions that can tolerate more asparaginase doses.6
Two main pathways of allergic reaction have been described for proteins7 : the classical pathway requires antigen-specific immunoglobulin E (IgE) antibodies, involves the FcεRI receptor, and mediates allergic reactions via the release of histamine. The alternative pathway requires high antigen-specific IgG antibody titers, involves the FcγRIII receptor, and mediates reactions by platelet-activating factor release. There is definite evidence supporting the role of the alternative pathway during asparaginase allergies 8 ; however, contribution by the classical pathway cannot be ruled out. Regardless of the mechanism of hypersensitivity, human leukocyte antigen (HLA) class II alleles play a critical role in these immune responses. Antibody production requires activation of CD4+ helper T cells, which involves the engagement of the T-cell receptor (TCR) with the HLA class II complex on antigen-presenting cells.9 Moreover, B cells process antigens, bind epitopes on the HLA class II protein, and present the HLA–antigen complex to primed helper T cells, resulting in cytokine secretion by T cells and differentiation of B cells into antibody-secreting plasma cells.10
Given the critical role HLA class II proteins play during adaptive immune responses and to the involvement of antigen-specific antibodies during hypersensitivity reactions, our primary goal was to investigate associations of HLA class II alleles with hypersensitivity reactions to asparaginase. An additional objective was to elucidate the mechanism of asparaginase-induced allergies by determining how polymorphic HLA amino acid variants alter the binding pocket of the HLA protein and affect asparaginase antigenicity.
Materials and methods
Patients
Five cohorts of patients were available to investigate the association of HLA class II alleles with asparaginase hypersensitivity in pediatric ALL patients (n = 3547; supplemental Figure 1). Informed consent from the parents or guardians and assent from the patients as appropriate were obtained according to institutional review board guidelines for treatment and for genomic research. Three cohorts were treated with St. Jude Children’s Research Hospital (SJCRH) protocols: Total XIIIA (n = 154, accrued patients from 1991 to 1994), Total XV (n = 498), and Total XVI (n = 271). Two cohorts were treated with Children’s Oncology Group (COG) protocols: POG 9906 (n = 234) and COG AALL0232 (n = 2390). Patients enrolled on SJCRH Total XIIIA, Total XV, and COG protocol POG 9906 received native E coli asparaginase (Elspar), whereas those enrolled on SJCRH Total XVI and COG AALL0232 protocols received PEGylated E coli asparaginase (Oncaspar) (Table 1). The combined cohort containing SJCRH and COG patients with imputed HLA class II alleles had a median age of 9.3 years at diagnosis, with a range from 0.71 to 30 years. The dose and frequency of asparaginase used varied by protocol and treatment arm (Table 1). Asparaginase hypersensitivities to Elspar or Oncaspar were graded according to the standard National Cancer Institute common toxicity criteria,11 with grade 2 hypersensitivity and higher considered as allergy. An enzyme-linked immunosorbent assay–based method was used to detect anti-E coli asparaginase IgG antibodies, as described previously.8,12 Patients were assigned a positive or negative status based on their titer value relative to the negative control process mean.8
Data sets used for HLA association study and for HLA class II allele imputation
| Data set . | Sample size . | % Allergy* . | Asparaginase formulation . | Asparaginase dose . | Genotyping platform . | SNPs used for HLA imputation . | Imputed SNPs . |
|---|---|---|---|---|---|---|---|
| T1DGC | 4 871 | NA | NA | NA | Illumina Immunochip | NA† | NA |
| Total XIIIA | 91 | 30.8 | Elspar (native E coli asparaginase) | 10 000 IU/m2, IM 3 times weekly (induction and reinduction phases; 18 doses) and on weeks 4, 8, 12, 16, 20, 24, and 28 of continuation | Illumina Exome Beadchip | 1 814 | 4 753 |
| Total XV | 320 | 45.6 | Elspar (native E coli asparaginase) | 10 000-25 000 IU/m2, IM 3 times weekly or once weekly (induction, continuation/reinduction; 24-28 doses) | Affymetrix 500K | 578 | 5 989 |
| Total XVI | 130 | 18.5 | Oncaspar (PEGylated E coli asparaginase) | 2 500-3 500 IU/m2, IV weekly (induction, continuation/reinduction; 5-17 doses) | Affymetrix 6.0 | 964 | 5 603 |
| POG 9906 | 125 | 28.0 | Elspar (native E coli asparaginase) | 6 000-15 000 IU/m2, IM 3 times weekly or once weekly (induction, consolidation, interim maintenance, delayed intensification; 52 doses) | Affymetrix 500K | 578 | 5 989 |
| AALL0232 | 1 204 | 11.4 | Oncaspar (PEGylated E coli asparaginase) | 2 500 IU/m2, IM weekly (induction, consolidation, interim maintenance, delayed intensification; 6-12 doses) | Affymetrix 6.0 | 964 | 5 603 |
| Data set . | Sample size . | % Allergy* . | Asparaginase formulation . | Asparaginase dose . | Genotyping platform . | SNPs used for HLA imputation . | Imputed SNPs . |
|---|---|---|---|---|---|---|---|
| T1DGC | 4 871 | NA | NA | NA | Illumina Immunochip | NA† | NA |
| Total XIIIA | 91 | 30.8 | Elspar (native E coli asparaginase) | 10 000 IU/m2, IM 3 times weekly (induction and reinduction phases; 18 doses) and on weeks 4, 8, 12, 16, 20, 24, and 28 of continuation | Illumina Exome Beadchip | 1 814 | 4 753 |
| Total XV | 320 | 45.6 | Elspar (native E coli asparaginase) | 10 000-25 000 IU/m2, IM 3 times weekly or once weekly (induction, continuation/reinduction; 24-28 doses) | Affymetrix 500K | 578 | 5 989 |
| Total XVI | 130 | 18.5 | Oncaspar (PEGylated E coli asparaginase) | 2 500-3 500 IU/m2, IV weekly (induction, continuation/reinduction; 5-17 doses) | Affymetrix 6.0 | 964 | 5 603 |
| POG 9906 | 125 | 28.0 | Elspar (native E coli asparaginase) | 6 000-15 000 IU/m2, IM 3 times weekly or once weekly (induction, consolidation, interim maintenance, delayed intensification; 52 doses) | Affymetrix 500K | 578 | 5 989 |
| AALL0232 | 1 204 | 11.4 | Oncaspar (PEGylated E coli asparaginase) | 2 500 IU/m2, IM weekly (induction, consolidation, interim maintenance, delayed intensification; 6-12 doses) | Affymetrix 6.0 | 964 | 5 603 |
IM, intramuscular; NA, not available.
% Allergy is the percentage of patients with hypersensitivities to asparaginase within each data set.
A total of 7792 SNPs were available from the T1DGC data set for SNP and HLA imputation.
Genotyping
Germline DNA collected after remission was interrogated using the Affymetrix Human Mapping 500K Array Set, the Affymetrix Genome-Wide Human single nucleotide polymorphism (SNP) Array 6.0, or the Illumina Exome Beadchip array (n = 3146; supplemental Figure 1).13 Ancestry was estimated using STRUCTURE on the basis of the ancestry-informative SNPs interrogated by each genotyping platform, as described previously.14
Imputation of HLA class II alleles and amino acid variants
To impute the HLA alleles of pediatric ALL patients using genome-wide SNP data, the Type 1 Diabetes Genetics Consortium (T1DGC) reference panel was used, as previously described.15 The T1DGC reference panel includes 4871 unrelated individuals of European ancestry, each with experimentally determined HLA-DRB1 and HLA-DQB1 alleles as well as genotypes interrogated by the Illumina Immunochip array. The SNP data required for HLA allele imputation included 7792 SNPs spanning hg18 chr6: 29 600 542 to 33 268 403 from the Immunochip array and single nucleotide variants from HLA-DRB1 and HLA-DQB1 genes inferred with the EMBL-EBI Immunogenetics HLA database (http://www.ebi.ac.uk/ipd/imgt/hla/).16 HLA allele imputations in ALL patients were restricted to those with ≥90% European ancestry because of the poor imputation accuracies attained in non-European individuals when imputing HLA alleles using a European reference panel.17-20 No SNP data within HLA-DPB1 were available in the T1DGC reference panel. The overlap of Illumina Immunochip SNPs from the T1DGC reference panel and SNPs typed in patient samples was determined (Table 1), and Immunochip SNPs that were not genotyped in patients were imputed using the BEAGLE software package21 with the T1DGC reference panel. The T1DGC data set and BEAGLE were used to impute the HLA-DRB1 and HLA-DQB1 alleles in ALL patients. HLA-DRB1 amino acid sequences were inferred using the imputed HLA-DRB1 alleles and the EMBL-EBI Immunogenetics HLA database.
HLA typing and determination of HLA-DRB1 and HLA-DQB1 imputation concordance
HLA testing was performed by molecular methods in a subset of patients (61 SJCRH patients). All but the earliest 3 samples were tested by a minimum of 2 of 3 methods: sequence-specific primers (Life Technologies), Luminex sequence-specific oligoprobes (LabType, One Lambda), and DNA sequencing (Abbott Molecular). The 3 earliest samples included in this study were tested only by sequence-specific primers for exon 2 of the HLA-DRB1 locus (Life Technologies, Carlsbad, CA).22 Extracted DNA was amplified using Taq polymerase and 95 primer pairs specific for areas of known variation in exon 2 of the HLA-DRB1 or HLA-DQB1 loci. Samples tested by Luminex sequence-specific oligoprobes were amplified by locus-specific biotinylated primers for exon 2 of the HLA-DRB1 or HLA-DQB1 loci. Oligonucleotide probes specific for known areas of variation were bound to specific beads of a given fluorescent dye mixture and analyzed in a Luminex flow cytometer. DNA sequencing was performed for HLA-DRB1 and HLA-DQB1 as described, with amplification performed using forward and reverse Big Dye–labeled primers for exon 2 of HLA-DRB1 and HLA-DQB1. Sequencing was performed on an ABI 3130xL or 3500xL genetic analyzer and sequences were evaluated with HLA allele analysis software from Conexio Genomics.23 The concordance between the experimentally determined HLA alleles and the imputed HLA alleles was assessed by determining whether the methods gave identical results for each patient within the subset.
Binding affinity
To estimate the binding affinity of unique HLA-DRB1 alleles for various asparaginase epitopes, the Immune Epitope Database (IEDB, www.iedb.org) was used.24 The protein sequence of native E coli asparaginase (Entrez Protein database, accession number P00805) without the signal sequence was parsed into overlapping 15-mer peptide fragments and each epitope was scored for binding to HLA-DRB1 (percentile rank) using a consensus, sequence-based method.25,26 A lower score indicates high-affinity binding; higher scores indicate low-affinity binding. The lowest score for each allele was determined, and the 50th percentile binding score of all HLA-DRB1 alleles was estimated. Alleles above the 50th percentile score were dichotomized into a “low-binding” category, and alleles below the 50th percentile score were dichotomized into a “high-binding” category.
Structural modeling and simulations
Homology models of HLA-DRB1*07:01, *14:01, *15:01, and *01:01 protein structures were made using the SwissModel server27-29 and 3PDO from the Research Collaboratory for Structural Bioinformatics protein data bank30 as the template. Electrostatic and polar interactions of key residues with their neighboring residues were calculated with 6Å cutoff using Schrodinger’s PyMol molecular visualization system. The peptide selected for docking into all 4 proteins was 212PKVGIVYNYANASDLPAKA231, which was predicted by the IEDB server to contain the region of highest binding affinity for the *07:01 and *14:01 alleles. ZDOCK31 was used to find initial dockings for each protein, and the docking with the lowest estimated binding free energy across the different registers was chosen. The dockings were refined using Rosetta FlexPepDock32 to place side chains and adjust the backbone pose within the starting register. A 20-ns molecular dynamics run with explicit water was performed using Amber12 with the ff99SB force field33,34 after minimization and equilibration. Binding free energy was estimated from snapshots of the simulation using the Molecular Mechanics/Generalized Born Surface Area method.35
Data analysis
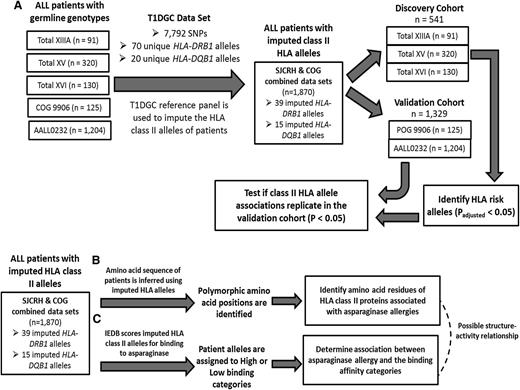
To evaluate the association between HLA-DRB1 and HLA-DQB1 alleles and asparaginase hypersensitivity, patients treated with SJCRH Total protocols (n = 541, Total XIIIA, Total XV, and Total XVI) were assigned to the discovery cohort and patients treated with COG protocols (n = 1329; POG 9906 and COG AALL0232) were assigned to the validation cohort. To determine the association between polymorphic HLA-DRB1 amino acid position or predicted binding-affinity categories with asparaginase hypersensitivity, the combined cohort was used (n = 1870). A general linear model was used to analyze the association of HLA alleles, polymorphic amino acid positions, and predicted binding-affinity categories with asparaginase hypersensitivity. Gender, age (<10 or ≥10 years), treatment arm, and ALL immunophenotype were included in the model as categorical covariates. Homozygous variant HLA-DRB1*07:01 genotypes and homozygous variant HLA-DRB1 amino acid residue groups with fewer than 5 observed patients were combined with the heterozygous variant groups where indicated. To account for multiple testing, P values were determined by randomly permuting HLA genotypes or amino acids 10 000 times, and P values were computed that reflected the probability of observing each result by chance (indicated as Padjusted). Differences between the binding free energy of the HLA-DRB1*07:01 protein and the free energy of HLA-DRB1*01:01, HLA-DRB1*14:01, and HLA-DRB1*15:01 were determined using a pairwise Wilcoxon rank sum test on the sets of snapshot binding energies collected from the Molecular Mechanics/Generalized Born Surface Area procedure. Permuted P values (Padjusted) < .05 were considered significant, and within our validation cohort, P values < .05 were considered significant. R statistical software (version 2.13.2) was used for analysis.
Results
Class II HLA alleles were imputed in 1870 patients of European descent that had data available on asparaginase hypersensitivity (Figure 1A; supplemental Figure 1; supplemental Table 1). Univariate analysis of potential risk factors for asparaginase hypersensitivity identified a higher risk for male patients (22% vs 17%; supplemental Table 2), age <10 years (23% vs 17%; supplemental Table 2), and for treatment with native E coli asparaginase rather than PEGylated asparaginase (39% vs 12%; supplemental Table 2). The incidence of asparaginase allergy varied by treatment protocol (P < 1.0 × 10−6; supplemental Table 2), ranging from 11% for COG ALL0232 to 46% for the SJCRH Total XV protocol. A multivariate analysis containing all covariates showed that gender, ALL immunophenotype, and treatment protocol were associated with asparaginase hypersensitivity (supplemental Table 2). T-lineage leukemia patients were only enrolled on SJCRH protocols Total XIIIA, Total XV, and Total XVI; within these protocols, we observed a higher risk of hypersensitivity among B-lineage leukemia patients compared with T-lineage leukemia patients (P = .01; supplemental Table 2). As a result, immunophenotype was included in the multivariate analysis even though it was not significant in all trials. Data on the phase of therapy in which the reaction occurred were available for 314 of the 370 cases. Within these patients, 85% of the reactions occurred during reexposure to asparaginase given in a phase after induction, suggesting that sensitization occurs during induction therapy in most cases.
HLA-DRB1 and HLA-DQB1 alleles were imputed for asparaginase hypersensitivity studies. The HLA-DRB1 and HLA-DQB1 alleles of patients enrolled on either SJCRH or COG protocols were imputed to 4-digit resolution using SNP data from the patients and the T1DGC reference panel with both SNP data and 4-digit resolution HLA typing. (A) The discovery cohort consisting of SJCRH patients was used to identify associations between asparaginase hypersensitivity reactions and imputed HLA-DRB1 and/or HLA-DQB1 alleles. The validation cohort consisted of COG patients. (B) Using the imputed HLA-DRB1 alleles, the HLA-DRB1 amino acid sequence was inferred for every patient, and the association between polymorphic amino acids and asparaginase hypersensitivity reactions was determined. (C) Using the IEDB, imputed HLA alleles were scored for their binding affinity to E coli asparaginase peptide fragments. The imputed HLA alleles of patients were categorized into “high” or “low” binding categories for asparaginase, and the categories were tested for association with asparaginase hypersensitivity.
HLA-DRB1 and HLA-DQB1 alleles were imputed for asparaginase hypersensitivity studies. The HLA-DRB1 and HLA-DQB1 alleles of patients enrolled on either SJCRH or COG protocols were imputed to 4-digit resolution using SNP data from the patients and the T1DGC reference panel with both SNP data and 4-digit resolution HLA typing. (A) The discovery cohort consisting of SJCRH patients was used to identify associations between asparaginase hypersensitivity reactions and imputed HLA-DRB1 and/or HLA-DQB1 alleles. The validation cohort consisted of COG patients. (B) Using the imputed HLA-DRB1 alleles, the HLA-DRB1 amino acid sequence was inferred for every patient, and the association between polymorphic amino acids and asparaginase hypersensitivity reactions was determined. (C) Using the IEDB, imputed HLA alleles were scored for their binding affinity to E coli asparaginase peptide fragments. The imputed HLA alleles of patients were categorized into “high” or “low” binding categories for asparaginase, and the categories were tested for association with asparaginase hypersensitivity.
Among the 1870 patients with leukemia, 39 unique HLA-DRB1 and 15 unique HLA-DQB1 alleles were imputed (Figure 2A-B). The overall concordance of HLA class II imputations was 91% for HLA-DRB1 and 97% for HLA-DQB1 in the subset of 61 patients with experimentally determined HLA types (supplemental Tables 3 and 4). For HLA-DRB1, 17 of 24 alleles had an imputation accuracy of 100% (supplemental Table 3), and for HLA-DQB1, 9 of 12 alleles had an imputation accuracy of 100% (supplemental Table 4).
HLA class II allele imputations resulted in 54 unique class II HLA alleles in patients. The imputation of HLA-DRB1 alleles within the combined data sets (n = 1870) resulted in (A) 39 unique HLA-DRB1 alleles and (B) 15 unique HLA-DQB1 alleles of varying allele frequency.
HLA class II allele imputations resulted in 54 unique class II HLA alleles in patients. The imputation of HLA-DRB1 alleles within the combined data sets (n = 1870) resulted in (A) 39 unique HLA-DRB1 alleles and (B) 15 unique HLA-DQB1 alleles of varying allele frequency.
In the discovery cohort, we identified an association between HLA-DRB1*07:01 and asparaginase hypersensitivity (P = .001 [Padjusted = .023], odds ratio [OR] = 1.92, 95% confidence interval [CI] = 1.29-2.87; Figure 3A-B). The association of HLA-DRB1*07:01 with asparaginase hypersensitivity was replicated in the validation cohort (Figure 3C; P = .014, OR = 1.49, CI = 1.08-2.03), and a higher frequency of hypersensitivity was detected in patients with the HLA-DRB1*07:01 allele across all 5 cohorts (supplemental Figure 2). Combining all SJCRH and COG patients, those carrying the risk allele had a 45% higher incidence of hypersensitivity compared with patients who did not (25.8% incidence of allergy for those carrying the risk allele vs a 17.8% incidence for those without the allele, P = 7.5 × 10−5 [Padjusted = .002], OR = 1.64, CI = 1.28-2.09; Figure 3D, supplemental Figure 3, supplemental Table 5). Furthermore, when patients were divided into the 536 patients who received native E coli asparaginase and the 1334 patients who received PEGylated E coli asparaginase (Table 1), HLA-DRB1*07:01 remained the strongest HLA allele associated with hypersensitivity in each group (P = .007 and P = .005, respectively).
Association of HLA-DRB1 and HLA-DQB1 alleles with asparaginase hypersensitivity. Using a general linear model adjusted for gender, age, treatment arm, and ALL immunophenotype, (A) HLA-DRB1*07:01 (P = .001 [Padjusted = .023]) was associated with asparaginase hypersensitivities within the discovery cohort (n = 541). The dashed horizontal red line identifies a permuted significance threshold of P < .05 that is adjusted for multiple testing. Patients with the HLA-DRB1*07:01 allele had a higher incidence of hypersensitivity compared with patients that did not have the allele within the (B) discovery cohort (n = 541), (C) validation cohort (n = 1329), and (D) the combined cohort (n = 1870).
Association of HLA-DRB1 and HLA-DQB1 alleles with asparaginase hypersensitivity. Using a general linear model adjusted for gender, age, treatment arm, and ALL immunophenotype, (A) HLA-DRB1*07:01 (P = .001 [Padjusted = .023]) was associated with asparaginase hypersensitivities within the discovery cohort (n = 541). The dashed horizontal red line identifies a permuted significance threshold of P < .05 that is adjusted for multiple testing. Patients with the HLA-DRB1*07:01 allele had a higher incidence of hypersensitivity compared with patients that did not have the allele within the (B) discovery cohort (n = 541), (C) validation cohort (n = 1329), and (D) the combined cohort (n = 1870).
To determine the protein sequence features of HLA-DRB1*07:01 that confer a greater risk of asparaginase hypersensitivity (Figure 1B), polymorphic amino acid variants of the HLA-DRB1 protein were inferred. A total of 30 polymorphic amino acid positions were identified within our patient population, with 16 positions having 3 or more amino acid residues possible (90 total residues within the 30 polymorphic amino acid positions).
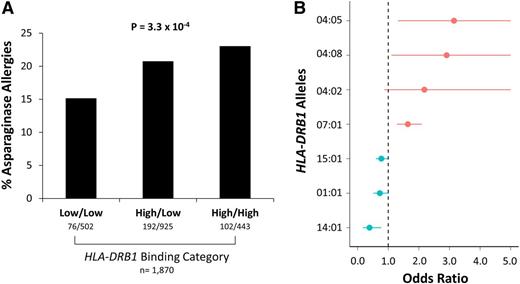
There were 10 risk amino acid positions that were associated with asparaginase hypersensitivity: a glycine at position 73 had the strongest association with asparaginase hypersensitivity (P = 6.45 × 10−6 [Padjusted < 1.00 × 10−4], OR = 1.76, CI = 1.38-2.26; Figure 4A-B, Table 2), 6 positions (11, 13, 14, 25, 30, and 74) were uniquely coded by HLA-DRB1*07:01 (P = 1.34 × 10−4 [Padjusted = .003], OR = 1.69, CI = 1.29-2.21; Figure 4A, Table 2), and 3 positions (57, 60, and 78) were associated with hypersensitivity to a lesser extent (P < 2.2 × 10−3 [Padjusted < .05]; Figure 4A, Table 2). Examination of the published crystal structure (PDB code 3PDO) of HLA-DR130 shows that positions 11, 13, 14, 25, and 30 of the HLA-DRB1 chain are all located within the β-sheet floor of the protein and positions 57, 60, 73, 74, and 78 are on the adjacent α helix in close proximity to positions 11 and 13 (Figure 5). The side chains of floor residues 11, 13, and 30 point toward the binding pocket, whereas those of residues 14 and 25 point toward the β-sheet floor. The amino acid residues at these positions associated with a higher risk of hypersensitivity (Table 2) were all present in the HLA-DRB1*07:01 allele. These results suggest that HLA-DRB1*07:01 may predispose to a higher incidence of allergies because structural features of the binding pocket may affect the interaction of asparaginase epitopes with the HLA-DRB1 protein. Two other HLA-DRB1 alleles within our data set contain a glycine at position 73 (Table 2, HLA-DRB1*03:01 and HLA-DRB1*03:02); both had an OR > 1.0, although neither allele reached statistical significance for association with allergy (supplemental Table 5). After adjusting for position 73 in the general linear model, only a tyrosine at position 37 was associated with hypersensitivity reactions (P = .002 [Padjusted = .036], OR = 2.02, CI = 1.56-2.63). This risk residue in position 37 was coded by 17 different HLA-DRB1 alleles in our data set, and the position was not associated with hypersensitivity without adjusting for the strongest association.
Polymorphic amino acid positions of HLA-DRB1 are associated with asparaginase hypersensitivity. (A) Amino acid position 73 had the strongest association with asparaginase hypersensitivity, and positions 11, 13, 14, 25, 30, 57, 60, 74, and 78 were associated with hypersensitivity to a lesser extent. The y-axis shows the permuted P value (Padjusted) of each association tested and the x-axis represents the amino acid positions of the HLA-DRB1 protein, excluding the leader signal sequence. The dashed horizontal red line identifies a permuted significance threshold of P < .05 that is adjusted for multiple testing. (B) Patients with a glycine (Gly) residue at HLA-DRB1 amino acid position 73 had a higher incidence of hypersensitivity compared with patients with an alanine (Ala/Ala) (n = 1,870, P = 6.45 × 10−6 [Padjusted < 1.00 × 10−4]).
Polymorphic amino acid positions of HLA-DRB1 are associated with asparaginase hypersensitivity. (A) Amino acid position 73 had the strongest association with asparaginase hypersensitivity, and positions 11, 13, 14, 25, 30, 57, 60, 74, and 78 were associated with hypersensitivity to a lesser extent. The y-axis shows the permuted P value (Padjusted) of each association tested and the x-axis represents the amino acid positions of the HLA-DRB1 protein, excluding the leader signal sequence. The dashed horizontal red line identifies a permuted significance threshold of P < .05 that is adjusted for multiple testing. (B) Patients with a glycine (Gly) residue at HLA-DRB1 amino acid position 73 had a higher incidence of hypersensitivity compared with patients with an alanine (Ala/Ala) (n = 1,870, P = 6.45 × 10−6 [Padjusted < 1.00 × 10−4]).
HLA-DRB1 polymorphic amino acid positions associated with asparaginase hypersensitivity
| AA position (no leader)* . | Risk residue† . | P value (Padjusted)‡ . | OR‡ . | 95% CI . | Other possible amino acids¶ . | HLA-DRB1 alleles with risk residue . |
|---|---|---|---|---|---|---|
| 11 | G | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | D, G, L, P, S, V | *07:01 |
| 13 | Y | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | F, G, H, R, S, Y | *07:01 |
| 14 | K | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | E, K | *07:01 |
| 25 | Q | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | Q,R | *07:01 |
| 30 | L | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | C, G, H, L, R, Y | *07:01 |
| 57 | V | 1.19 × 10−3 (.023) | 1.53 | 1.18-1.98 | A, D, S | *07:01, *09:01, *12:01 |
| 60 | S | 1.19 × 10−3 (.023) | 1.53 | 1.18-1.98 | H, Y | *07:01, *09:01, *12:01 |
| 73 | G | 6.45 × 10−6 (<1.00 × 10−4) | 1.76 | 1.38-2.26 | A, G | *07:01,*03:01, *03:02 |
| 74 | Q | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | A, E, L, Q, R | *07:01 |
| 78 | V | 2.18 × 10−3 (.042) | 1.51 | 1.16-1.97 | V, Y | *07:01, *09:01 |
| AA position (no leader)* . | Risk residue† . | P value (Padjusted)‡ . | OR‡ . | 95% CI . | Other possible amino acids¶ . | HLA-DRB1 alleles with risk residue . |
|---|---|---|---|---|---|---|
| 11 | G | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | D, G, L, P, S, V | *07:01 |
| 13 | Y | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | F, G, H, R, S, Y | *07:01 |
| 14 | K | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | E, K | *07:01 |
| 25 | Q | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | Q,R | *07:01 |
| 30 | L | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | C, G, H, L, R, Y | *07:01 |
| 57 | V | 1.19 × 10−3 (.023) | 1.53 | 1.18-1.98 | A, D, S | *07:01, *09:01, *12:01 |
| 60 | S | 1.19 × 10−3 (.023) | 1.53 | 1.18-1.98 | H, Y | *07:01, *09:01, *12:01 |
| 73 | G | 6.45 × 10−6 (<1.00 × 10−4) | 1.76 | 1.38-2.26 | A, G | *07:01,*03:01, *03:02 |
| 74 | Q | 1.34 × 10−4 (.003) | 1.69 | 1.29-2.21 | A, E, L, Q, R | *07:01 |
| 78 | V | 2.18 × 10−3 (.042) | 1.51 | 1.16-1.97 | V, Y | *07:01, *09:01 |
AA position is the amino acid position of the HLA-DRB1 protein sequence without the leader signal sequence.
Risk residue is the amino acid associated with asparaginase hypersensitivity.
For the association between the risk residue and asparaginase hypersensitivity, Padjusted values were determined by permutation to account for multiple testing.
Other possible amino acids refer to other amino acids within that amino acid position observed in our patients (n = 1870).
The 3-dimensional structure of HLA-DR. The 3-dimensional ribbon model of HLA-DR is based on Protein Data Bank entry 3PDO with the DR α chain shown in violet and the DR β chain in gray. Amino acid positions identified within the HLA-DRB1 protein by the association analysis are shown for positions 11 (black), 13 (blue), 14 (cyan), 25 (orange), 30 (brown), 57 (yellow), 60 (pink), 73 (red), 74 (green), and 78 (purple).
The 3-dimensional structure of HLA-DR. The 3-dimensional ribbon model of HLA-DR is based on Protein Data Bank entry 3PDO with the DR α chain shown in violet and the DR β chain in gray. Amino acid positions identified within the HLA-DRB1 protein by the association analysis are shown for positions 11 (black), 13 (blue), 14 (cyan), 25 (orange), 30 (brown), 57 (yellow), 60 (pink), 73 (red), 74 (green), and 78 (purple).
The binding affinities of HLA-DRB1 alleles for asparaginase epitopes were estimated using the IEDB, which uses a consensus sequence-based method to estimate binding affinities25,26 (Figure 1C; supplemental Figure 4). Patients with 1 or 2 high-binding alleles had a higher incidence of asparaginase hypersensitivities compared with patients with 2 low-binding alleles (P = 3.3 × 10−4, OR = 1.38, CI = 1.16-1.64; Figure 6A). HLA-DRB1*07:01 was a predicted high-binding allele (supplemental Table 5), consistent with its association with asparaginase hypersensitivity reactions, and all HLA-DRB1 alleles with significant (P < .1) association with allergies (supplemental Table 5) had predicted binding affinities consistent with their estimated OR (Figure 6B), including 3 alleles coding for a tyrosine at position 37 (HLA-DRB1*04:05, HLA-DRB1*04:08, and HLA-DRB1*04:02).
High-affinity binding of HLA-DRB1 alleles was associated with asparaginase hypersensitivity reactions. (A) Patients with high-affinity binding alleles had a higher incidence of asparaginase hypersensitivity compared with patients with low-affinity binding alleles (n = 1870, P = 3.3 × 10−4). (B) Estimated ORs for association of HLA-DRB1*01:01, HLA-DRB1*04:02, HLA-DRB1*04:05, HLA-DRB1*04:08, HLA-DRB1*07:01, HLA-DRB1*14:01, and HLA-DRB1*15:01 alleles with asparaginase hypersensitivity, all at P < .1, are shown vs the predicted binding category (predicted low-binding alleles are shown in blue; predicted high-binding alleles are shown in red). All HLA-DRB1 alleles had predicted binding affinities consistent with the estimated odds ratios.
High-affinity binding of HLA-DRB1 alleles was associated with asparaginase hypersensitivity reactions. (A) Patients with high-affinity binding alleles had a higher incidence of asparaginase hypersensitivity compared with patients with low-affinity binding alleles (n = 1870, P = 3.3 × 10−4). (B) Estimated ORs for association of HLA-DRB1*01:01, HLA-DRB1*04:02, HLA-DRB1*04:05, HLA-DRB1*04:08, HLA-DRB1*07:01, HLA-DRB1*14:01, and HLA-DRB1*15:01 alleles with asparaginase hypersensitivity, all at P < .1, are shown vs the predicted binding category (predicted low-binding alleles are shown in blue; predicted high-binding alleles are shown in red). All HLA-DRB1 alleles had predicted binding affinities consistent with the estimated odds ratios.
Homology models were made of the high-affinity HLA-DRB1*07:01 protein and of 3 proteins predicted to have low-binding affinity (HLA-DRB1*14:01, *15:01, and *01:01). All predicted low-binding alleles had OR < 1 for their association with allergy, and they showed the strongest association to protect against asparaginase hypersensitivity (supplemental Table 5; Figure 6B). The amino acid residues at positions 11, 13, 14, 25, 30, 57, 60, 73, 74, and 78 for these 3 low-affinity binding alleles differ from the corresponding amino acid sequence of HLA-DRB1*07:01 (supplemental Table 6). Structural comparison of these 4 alleles shows that several salt bridges and/or polar contacts exist between amino acids in risk positions and neighboring residues for the 3 low-affinity binding alleles, but not for the HLA-DRB1*07:01 structure (supplemental Table 6). This, together with the occurrence of glycine at positions 11 and 73 in the HLA-DRB1*07:01 protein, suggests that the HLA-DRB1*07:01 protein may have more flexibility in the peptide-binding groove compared with the HLA-DRB1*14:01, *15:01, and *01:01 proteins, which may result in higher affinity binding between asparaginase epitopes and the HLA-DR structure.
To complement the results generated using the IEDB, we used a molecular dynamics–based method to provide a structure-based estimate of the binding free energy between the HLA protein and the putatively most immunogenic peptide region of asparaginase. For each of the 4 HLA alleles for which homology models were built, we first applied a docking procedure that aligned the peptide in several different registers within the binding groove of the HLA model. The molecular dynamics procedure was then applied for each register of each allele, and the register with the lowest binding free energy for each allele was compared. The HLA-DRB1*07:01 allele was estimated to bind the asparaginase peptide with significantly lower binding free energy (higher affinity) than the other 3 alleles (supplemental Figure 5, P < 1.0 × 10−6).
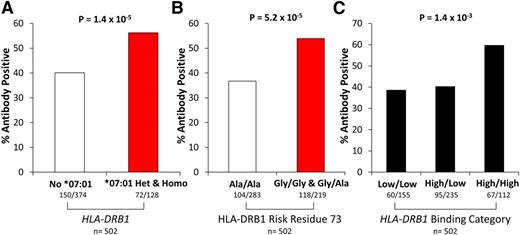
Anti-asparaginase IgG antibody data were available in 502 SJCRH patients (89 treated in Total XIIIA, 283 in Total XV, and 130 in Total XVI). A higher incidence of anti-asparaginase IgG antibody positive patients was detected in carriers of the HLA-DRB1*07:01 allele (Figure 7A, P = 1.4 × 10−5, OR = 2.92, CI = 1.82-4.80), in patients with a glycine risk residue at amino acid position 73 (Figure 7B, P = 5.2 × 10−5, OR = 2.39, CI = 1.57-3.67), or in patients with 2 high-affinity binding alleles (Figure 7C, P = 1.4 × 10−3, OR = 1.59, CI = 1.20-2.12) compared with patients with one or 2 low-affinity binding alleles, consistent with the allergy data. The proportion of HLA-DRB1*07:01 allele carriers among allergy cases that tested positive for anti-asparaginase IgG antibodies (n = 141) vs those who were negative (n = 81) was 35% vs 28% (P = .37), although power to detect any difference was low because of the low sample size.
Asparaginase antibodies were associated with HLA-DRB1*07:01 genotype, amino acid composition, and binding affinity. Anti-asparaginase IgG antibody status was available for 502 ALL patients. A higher incidence of asparaginase antibodies was detected for patients with (A) HLA-DRB1*07:01 (P = 1.4 × 10−5), (B) a glycine amino acid within HLA-DRB1 amino acid position 73 (P = 5.2 × 10−5), or (C) HLA-DRB1 alleles with predicted high-affinity binding for asparaginase (P = 1.4 × 10−3). Ala, alanine; Gly, glycine; het, heterozygous; homo, homozygous.
Asparaginase antibodies were associated with HLA-DRB1*07:01 genotype, amino acid composition, and binding affinity. Anti-asparaginase IgG antibody status was available for 502 ALL patients. A higher incidence of asparaginase antibodies was detected for patients with (A) HLA-DRB1*07:01 (P = 1.4 × 10−5), (B) a glycine amino acid within HLA-DRB1 amino acid position 73 (P = 5.2 × 10−5), or (C) HLA-DRB1 alleles with predicted high-affinity binding for asparaginase (P = 1.4 × 10−3). Ala, alanine; Gly, glycine; het, heterozygous; homo, homozygous.
Discussion
Patients with low asparaginase exposure during treatment of malignancy have been reported to have increased risk of relapse.6 Patients who develop hypersensitivity reactions or neutralizing antibodies to asparaginase have been shown to have increased asparaginase clearance leading to subtherapeutic serum concentrations of the drug.2,36-44 The frequency of asparaginase hypersensitivity has been associated with the pharmaceutical preparation of asparaginase,6,45 tumor immunophenotype,8,46 the treatment schedule of asparaginase,47 and racial ancestry48 ; importantly, we controlled for these covariates in our analyses of whether HLA genotypes were related to allergy in these prospective clinical trials. The influence of HLA genes on asparaginase hypersensitivity has not been studied previously. Herein, we identified that there is an association of the HLA-DRB1*07:01 allele with asparaginase hypersensitivity and anti-asparaginase antibodies (Figure 3A-D; Figure 7A). Nevertheless, a limitation of our study could be heterogeneity in reporting clinical allergy to asparaginase. Although it is likely that the most important cases of first instance of clinical allergy to asparaginase are captured by allergies of grade 2 and higher because of severity of those reactions (eg, rash, flushing, urticaria, dyspnea, drug fever ≥38°C), and these cases often precipitate therapeutic intervention for future doses, it is also possible that clinicians may have used antihistamines or glucocorticoids that attenuated or minimized reactions to grade 1 or lower, and thus some reactions may have been missed in our study.
Variants in HLA genes on chromosome 6 is linked with more autoimmune diseases than any other genomic region.49,50 Genetic variants within HLA genes have been associated with several diseases including celiac disease,51 rheumatoid arthritis,52,53 type I diabetes,54,55 and multiple sclerosis.56 Several HLA-B alleles have been associated with small molecule drug allergy, including abacavir,57,58 and adverse reactions to carbamazepine59 and allopurinol.60-62 HLA allele associations with immune responses to protein-based pharmaceuticals have also been reported for interferon-β (HLA-DRB1*04:01 and *04:08)63,64 and for recombinant human erythropoietin (HLA-DRB1*09).65,66 Furthermore, HLA-DRB1*07:01 has previously been linked to elevated levels of serum alanine aminotransferase during ximelagatran and lapatinib treatment through a drug-induced adaptive immune response.9,22
Although direct experimental HLA typing is costly and laborious, imputation of HLA types using SNP genotyping arrays has been shown to provide a reasonably accurate and cost-effective way of investigating drug hypersensitivity or disease association with classical HLA alleles.15,52,67,68 The HLA-DRB1 and HLA-DQB1 allele imputation accuracies for individuals of European descent have ranged from 72% to 98%.17,18,20,52 We achieved similar imputation accuracies of 91% and 97% for HLA-DRB1 and HLA-DQB1, respectively; furthermore, the imputation concordance for HLA-DRB1*07:01 was 100% (supplemental Table 3). We took advantage of imputation and found that the HLA-DRB1*07:01 allele was associated with allergy to asparaginase.
Inferring polymorphic HLA amino acid positions from HLA alleles69 allows for interrogation of specific residues within the HLA binding pocket for association with hypersensitivity. We identified HLA-DRB1 amino acid positions 11, 13, 14, 25, 30, 37, 57, 60, 73, 74, and 78 associated with asparaginase hypersensitivity. Interestingly, several of these have been associated with autoimmune disease: position 11 with sarcoidosis,70 positions 13 and 60 with multiple sclerosis,71,72 position 37 with psoriasis vulgaris,73 position 57 with Vogt-Koyanagi-Harada disease,74 position 78 with primary biliary cirrhosis,75 and position 74 with Graves’ disease76,77 and autoimmune polyglandular syndrome.78 Moreover, association of HLA-DRB1 amino acid positions 11, 13, and 74 with rheumatoid arthritis susceptibility was recently reported,52 suggesting that the HLA-DRB1 amino acid positions that predispose to asparaginase allergy may also be important for other immune responses.
Binding of epitopes to the HLA class II protein is required for antigen presentation to CD4+ T cells and to mediate an immune response.79 Epitope binding prediction tools have been used for vaccine development strategies,80 and for designing nonimmunogenic protein-based pharmaceuticals, such as factor VIII81 and asparaginase.82 Interestingly, the most immunogenic asparaginase region identified in our study using IEDB (212PKVGIVYNYANASDLPAKA231) was shown by Cantor et al82 to be the most immunogenic fragment of asparaginase using a T-cell activation assay and transgenic HLA-DRB1*04:01 mice. Whether less immunogenic formulations of asparaginase could be developed for specific HLA-DRB1 alleles that maintain undiminished asparaginase activity is unknown. No other studies have used human HLA class II loci to identify immunodominant epitopes of native E coli asparaginase. Using the IEDB, we found that patients harboring the HLA-DRB1 alleles with high-affinity binding to asparaginase epitopes had a higher incidence of asparaginase hypersensitivity compared with those with only low-affinity binding alleles (Figure 6A). Furthermore, HLA-DRB1*07:01 was predicted to have high-affinity binding for asparaginase epitopes (Figure 6B). Comparing HLA-DR structures with low-affinity binding alleles to the HLA-DR structure with the HLA-DRB1*07:01 allele (supplemental Table 6) suggests that inherited variation in HLA-DRB1 results in variation in amino acids within the binding pocket of the protein that can influence the interaction of epitopes with the HLA-DR protein and affect the immunogenicity of asparaginase. This is the first known report to use IEDB to computationally predict the immunogenicity of a drug based on patient HLA-DRB1 genotypes; such an approach might be useful for other protein-based drugs in the future.
In conclusion, we identified an association between the HLA-DRB1*07:01 allele and hypersensitivity reactions to asparaginase. Taken together with its association with anti-asparaginase antibodies, the polymorphic amino acids encoded, and structural analysis of the HLA-DR proteins, our results suggest a structure–activity relationship whereby HLA-DRB1 alleles that encode for polymorphic amino acid variants within the HLA-DRB1 protein that confer high-affinity binding to asparaginase epitopes can lead to a higher frequency of asparaginase hypersensitivity compared with HLA alleles that confer low-affinity binding. Using the IEDB and the protein sequence of the biologic drug, it is possible to a priori identify patients that are predisposed to developing an immune response. Strategies that can identify patients that are predisposed to developing allergic reactions to protein therapeutics may help lower frequency of immunogenicity during clinical trials by excluding high-risk patients from the study, selectively directing the use of premedications, or perhaps even by developing “designer therapeutics” as alternative forms of the protein to permit the use of HLA-specific preparations in predisposed individuals. Our results emphasize the importance of variation at the HLA locus for predicting the immunogenicity of protein therapeutics.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The study was supported by the National Cancer Institute (grants GM 92666, CA 21765, CA 142665, CA 36401, CA98543 [COG Chair's grant], CA98413 [COG Statistical Center], and CA114766 [COG Specimen Banking]) and the American Lebanese Syrian Associated Charities.
S.P.H. is the Ergen Family Chair in Pediatric Cancer.
Authorship
Contribution: C.A.F. and M.V.R. designed the project, analyzed the data, interpreted the data, and drafted the manuscript; C.A.F., C.S., and W.Y. performed statistical analysis and HLA imputations; M.D. and D.B. performed the structural analysis; V.T. performed the HLA typing; S.O.G., W.M.C., P.C., and S.S.R. provided the T1DGC data; E.L., W.P.B., M.L.L., E.A.R., N.J.W., S.P.H., W.L.C., S.J., C.H.P., W.E.E., and M.D. were investigators for the clinical protocols; C.L., L.B.R., T.C., and P.S. interpreted the data; and all authors contributed to the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mary V. Relling, St. Jude Children’s Research Hospital, 262 Danny Thomas Place, Memphis, TN 38105; e-mail: mary.relling@stjude.org.


![Figure 3. Association of HLA-DRB1 and HLA-DQB1 alleles with asparaginase hypersensitivity. Using a general linear model adjusted for gender, age, treatment arm, and ALL immunophenotype, (A) HLA-DRB1*07:01 (P = .001 [Padjusted = .023]) was associated with asparaginase hypersensitivities within the discovery cohort (n = 541). The dashed horizontal red line identifies a permuted significance threshold of P < .05 that is adjusted for multiple testing. Patients with the HLA-DRB1*07:01 allele had a higher incidence of hypersensitivity compared with patients that did not have the allele within the (B) discovery cohort (n = 541), (C) validation cohort (n = 1329), and (D) the combined cohort (n = 1870).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/8/10.1182_blood-2014-03-563742/4/m_1266f3.jpeg?Expires=1769094121&Signature=nk9cAoAFnjlr6BLYd9CZS3lzHpYwrdfQqIk8VpxRaK1MUFZOmemnizuE2ND-tGD2AZJvRFyt52ZAfjmGXPNZAoY9jKIdMlEipnPk04icJnLXUPkHJqa2vQM1~BLFOwb17~CXKns8lTHyexq2aJc91ozJiomDbkSikkTUuan~6b56FoFBBeK56c7ke0GKv5Dt3tyE0Q8jHziy5ez72Jw7LT9HjNy~HPkJt5DXz4HSbeCZ5fkdf0fFc53J0RLsbXliKQ8K05WjryVJ~Ql68G6dQ5b5pHGXEvGx06FE-U1vV7OcG2zOPx0ZCyhSCC9NPJTExNl~-cW10pUqtLRNdywcEA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. Polymorphic amino acid positions of HLA-DRB1 are associated with asparaginase hypersensitivity. (A) Amino acid position 73 had the strongest association with asparaginase hypersensitivity, and positions 11, 13, 14, 25, 30, 57, 60, 74, and 78 were associated with hypersensitivity to a lesser extent. The y-axis shows the permuted P value (Padjusted) of each association tested and the x-axis represents the amino acid positions of the HLA-DRB1 protein, excluding the leader signal sequence. The dashed horizontal red line identifies a permuted significance threshold of P < .05 that is adjusted for multiple testing. (B) Patients with a glycine (Gly) residue at HLA-DRB1 amino acid position 73 had a higher incidence of hypersensitivity compared with patients with an alanine (Ala/Ala) (n = 1,870, P = 6.45 × 10−6 [Padjusted < 1.00 × 10−4]).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/8/10.1182_blood-2014-03-563742/4/m_1266f4.jpeg?Expires=1769094121&Signature=goR22-s5ymsNN-sf6A9-MnMI~7aMz6XqtleLOdP67enRxsep68khZttKshymyDJ0tc0hAyY9olGsVKtPJZjbBzp1FRnwVrzwQ356BXT8hZeEgJDlr9Ah~QlXMtZqcPbbGniHQ5IBqgYCE5bmWzS8B04hZm-i6uLEFtjnzcEki~eiT3gjqaQ7mIhGZd5jf1MDaiYWZl8czomXDWq1lFdu1Y-5INpH5PdUpT~AXKKPrJwTtIDxWlNWHAAuRQVuqc~KsUoGy8XUFAwQrxgv5F9wjtRfKug1rXmYrwqcMnnjqS8tCxE9JUbWNE-rds2BXpyvVuGkZP6ej4b0SbarFexWNg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)