Key Points
IRF8 promotes Gata2 expression in GPs, thereby playing a key role in the development of basophils and mast cells.
Abstract
Basophils and mast cells play critical roles in host defense against pathogens and allergic disorders. However, the molecular mechanism by which these cells are generated is not completely understood. Here we demonstrate that interferon regulatory factor-8 (IRF8), a transcription factor essential for the development of several myeloid lineages, also regulates basophil and mast cell development. Irf8−/− mice displayed a severe reduction in basophil counts, which was accounted for by the absence of pre-basophil and mast cell progenitors (pre-BMPs). Although Irf8−/− mice retained peripheral tissue mast cells, remaining progenitors from Irf8−/− mice including granulocyte progenitors (GPs) were unable to efficiently generate either basophils or mast cells, indicating that IRF8 also contributes to the development of mast cells. IRF8 appeared to function at the GP stage, because IRF8 was expressed in GPs, but not in basophils, mast cells, and basophil/mast cell-restricted progenitor cells. Furthermore, we demonstrate that GATA2, a transcription factor known to promote basophil and mast cell differentiation, acts downstream of IRF8. These results shed light on the pathways and mechanism underlying the development of basophils and mast cells.
Introduction
Basophils and mast cells are key effector cells involved in protection against infection and allergic responses.1 Basophils are the rarest form of granulocytes, representing <1% of leukocytes in the peripheral blood, whereas mast cells are distributed throughout the mucosal and connective tissues.1,2 A representative function common to both basophils and mast cells is the release of chemical mediators in the basophilic granules once IgE-bound high-affinity IgE receptor (FcεRI) crosslinks with antigens. Nonredundant basophil roles have been revealed recently by in vivo analyses.2-4 These include protective T helper 2 (Th2) immune responses against helminthes via the secretion of IL-4 from basophils,5-7 Th2-dependent atopic dermatitis-like skin inflammation against haptens or peptide antigens via antigen presentation on basophil major histocompatibility complex II,8 and IgG-dependent systemic anaphylaxis via the secretion of platelet-activating factor from basophils.9 Novel roles for mast cells in immunity also have been demonstrated recently. Mast cells stimulate dendritic cell (DC) proliferation by secreting FMS-like tyrosine kinase 3 (Flt3) ligand during parasite infection, and are capable of presenting antigens to CD8+ T cells.10,11
The developmental pathway of basophils and mast cells has been actively studied in mice. Basophils and mast cells are generated from bone marrow hematopoietic stem cells by passing through common myeloid progenitors and granulocyte-monocyte progenitors (GMPs).12,13 Several downstream basophil and/or mast cell progenitor populations have been identified in the bone marrow. Granulocyte progenitors (GPs) generate granulocytes (ie, neutrophils, eosinophils, and basophils) and mast cells.14,15 GPs are comprised of 2 subpopulations (Flt3+ and Flt3−). Flt3− GPs have been shown to generate basophils more efficiently than Flt3+ GPs.15 Pre-basophil and mast cell progenitors (pre-BMPs) give rise to basophils and mast cells.16 Basophil progenitors (BaPs) and mast cell progenitors (MCPs), identified in the pioneer studies,17,18 are committed to differentiate into basophils and mast cells, respectively. In addition, basophil/mast cell progenitors (BMCPs) in the spleen have been reported as an intermediate between bone marrow GMPs and BaPs,17 but recent studies have shown that BMCPs give rise mostly to mast cells.15,16 It is noteworthy that GMPs are now thought to be a heterogeneous population containing monocyte–DC progenitors19 and the aforementioned pre-BMPs. The relationships between these progenitor populations have not been definitively established.
Cell differentiation is achieved through the coordinated regulation of gene expression, in which transcription factors play central roles. Several transcription factors, such as GATA-binding protein-1 (GATA1),20,21 GATA2,17,22 and signal transducer and activator of transcription-5 (STAT5),16,23 are known to be essential for the development of both basophils and mast cells. In addition, basophil differentiation depends on CCAAT/enhancer binding protein-α (C/EBPα)16,17,22 and runt-related transcription factor-115 ; mast cell differentiation requires microphthalmia-associated transcription factor (MITF).16 Interferon regulatory factor-8 (IRF8) is a transcription factor required for the generation of several immune cell types, especially within the myeloid cell lineage.24 IRF8 is highly expressed in the mononuclear phagocyte (DC and monocyte/macrophage) and B-cell lineages, but is absent in neutrophils.25-27 We have shown, along with other laboratories, that IRF8 is indispensable for the development of DCs, especially the plasmacytoid and CD8α+ DC subsets, and monocytes, especially the Ly6C+ inflammatory monocyte subset.28-31 Within the granulocyte lineage, IRF8 has been shown to inhibit neutrophil differentiation, whereas it promotes eosinophil differentiation.27,28,32
In this study, we identified a previously unrecognized role for IRF8 in the development of basophils and mast cells. Based on our results, we discuss the pathway and underlying mechanism of basophil and mast cell development.
Methods
Mice
Ly5.1, Ly5.2, Irf8−/−,33 Irf8Irf8Gfp/WT,26 Spib−/−,34 and KitW-sh/W-sh mice35 in a C57BL/6 background were used at 7 to 9 weeks of age. All the animal experiments were conducted in accordance with the Guidelines for Proper Conduct of Animal Experiments (Science Council of Japan), and all the protocols were approved by the institutional review boards of Yokohama City University (protocols #F11-85 and #F-A-14-070).
Other methods
Detailed methods describing cell isolation, flow cytometry, cell culture, toluidine blue staining, mast cell counting, generation of bone marrow chimeras, adoptive transfer of GPs, retroviral transduction, microarray analysis, reverse transcription-quantitative polymerase chain reaction (RT-qPCR), and in silico analysis of transcription factor binding motifs are available in the supplemental Methods on the Blood Web site. Microarray data are accessible from the Gene Expression Ominibus/National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/geo/) (accession #GSE54887).
Results
Analysis of basophil, mast cell, and progenitor counts in Irf8−/− mice
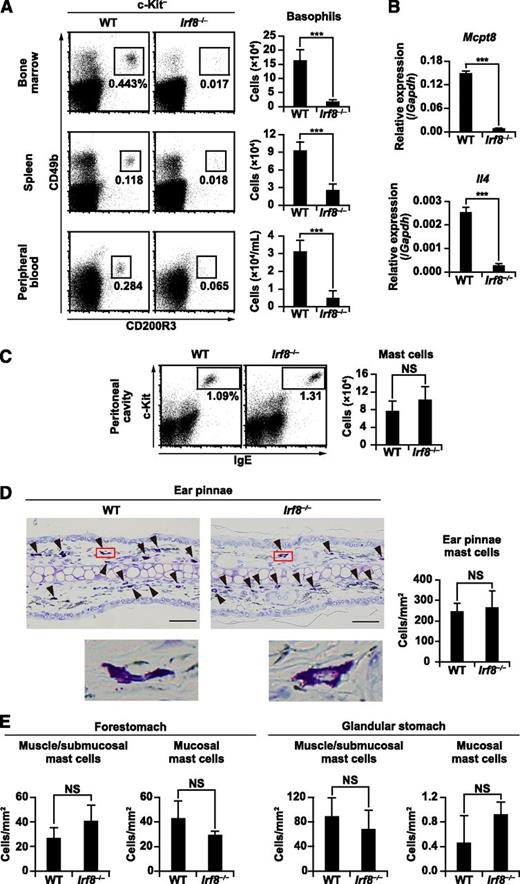
Basophil counts in the bone marrow, spleen, and peripheral blood of Irf8−/− mice were examined by flow cytometry. In all of these organs, both the percentages and absolute numbers of basophils, defined as c-Kit− CD49b+ CD200R3+ or c-Kit− CD49b+ IgE+ cells, were severely diminished in Irf8−/− mice when compared with wild-type (WT) mice (Figure 1A; supplemental Figure 1). RT-qPCR confirmed that the expression of the basophil-specific gene Mcpt8 and Il4 was impaired in Irf8−/− bone marrow (Figure 1B). In contrast, the generation and distribution of peripheral tissue mast cells, defined as c-Kit+ IgE+ cells in the peritoneal cavity (supplemental Methods and supplemental Figure 2) or toluidine blue–stained cells in the skin and gastrointestinal tract, were comparable in WT and Irf8−/− mice (Figures 1C-E).
Basophil and mast cell counts in Irf8−/− mice. (A,C) Flow cytometric analysis of basophils (A) and mast cells (C) in WT and Irf8−/− mice. Representative flow cytometry data are shown. Values in the bar graphs are means ± standard deviations from 5 to 7 mice of each genotype. (B) RT-qPCR analysis of Mcpt8 and Il4 in freshly isolated, unstimulated WT and Irf8−/− bone marrow cells. Values in the bar graph are means ± standard deviations from 3 mice of each genotype. (D) Representative toluidine blue–stained sections of ear pinnae (left). The arrowheads indicate typical mast cells. Insets show high magnification. Bars represent 50 μm (D). The mast cells in toluidine blue–stained sections were counted and normalized per unit area (mm2) of tissue examined (right). (E) Toluidine blue–stained sections of the stomach were counted as described in (D). Values in the bar graphs are presented as the mean ± standard deviation of sections derived from 3 mice of each genotype. ***P < .001 (Student t test). NS, not significant.
Basophil and mast cell counts in Irf8−/− mice. (A,C) Flow cytometric analysis of basophils (A) and mast cells (C) in WT and Irf8−/− mice. Representative flow cytometry data are shown. Values in the bar graphs are means ± standard deviations from 5 to 7 mice of each genotype. (B) RT-qPCR analysis of Mcpt8 and Il4 in freshly isolated, unstimulated WT and Irf8−/− bone marrow cells. Values in the bar graph are means ± standard deviations from 3 mice of each genotype. (D) Representative toluidine blue–stained sections of ear pinnae (left). The arrowheads indicate typical mast cells. Insets show high magnification. Bars represent 50 μm (D). The mast cells in toluidine blue–stained sections were counted and normalized per unit area (mm2) of tissue examined (right). (E) Toluidine blue–stained sections of the stomach were counted as described in (D). Values in the bar graphs are presented as the mean ± standard deviation of sections derived from 3 mice of each genotype. ***P < .001 (Student t test). NS, not significant.
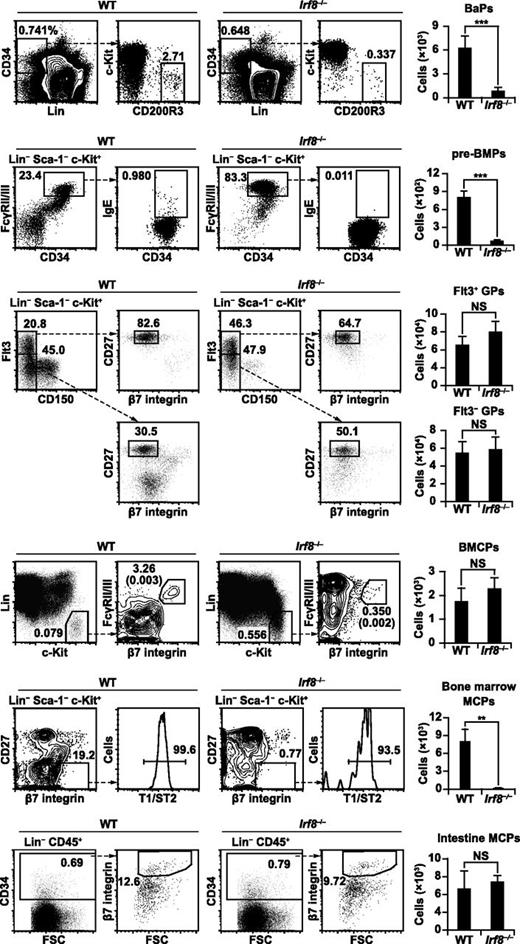
Next we examined the counts of progenitor populations, namely BaPs (lineage markers-negative [Lin−] CD34+ c-Kit− CD200R3+), pre-BMPs (Lin− Sca-1− c-Kit+ CD34+ FcγRII/III+ IgE+), and GPs (Lin− Sca-1− c-Kit+ CD150− β7 integrin− CD27+; Flt3+ or Flt3−) in the bone marrow and BMCPs (Lin− c-Kit+ FcγRII/III+ β7 integrin+) in the spleen (Figure 2). We found that the numbers of BaPs and pre-BMPs were significantly reduced in Irf8−/− mice, whereas the numbers of GPs (both Flt3+ and Flt3−) and BMCPs were unaffected. These results suggest that IRF8 is required for generating pre-BMPs.
Basophil and mast cell progenitor populations in Irf8−/− mice. Flow cytometric analysis of basophil/mast cell progenitor populations in WT and Irf8−/− mice. Representative flow cytometry data are shown. Numbers in the parentheses indicate the percentages relative to total cell counts. Values in the bar graphs are means ± standard deviations from 5 to 7 mice of each genotype. The reduced percentages of Lin− cKit+ FcγRII/III− CD34− cells (megakaryocyte-erythroid progenitors) and Lin− CD150+ Flt3− cells (megakaryocyte-erythroid progenitor-like population) in Irf8−/− mice are consistent with previous reports.37 **P < .01; ***P < .001 (Student t test). FSC, forward scatter; NS, not significant.
Basophil and mast cell progenitor populations in Irf8−/− mice. Flow cytometric analysis of basophil/mast cell progenitor populations in WT and Irf8−/− mice. Representative flow cytometry data are shown. Numbers in the parentheses indicate the percentages relative to total cell counts. Values in the bar graphs are means ± standard deviations from 5 to 7 mice of each genotype. The reduced percentages of Lin− cKit+ FcγRII/III− CD34− cells (megakaryocyte-erythroid progenitors) and Lin− CD150+ Flt3− cells (megakaryocyte-erythroid progenitor-like population) in Irf8−/− mice are consistent with previous reports.37 **P < .01; ***P < .001 (Student t test). FSC, forward scatter; NS, not significant.
The findings that the counts of pre-BMPs, but not BMCPs and mast cells, were severely diminished by IRF8 deficiency prompted us to examine MCPs in the bone marrow (Lin− Sca-1− c-Kit+ CD27− β7 integrin+ T1/ST2+) and peripheral tissues (Lin− CD45+ CD34+ β7 integrin+). MCPs, potentially derived from pre-BMPs and BMCPs, circulate in peripheral blood and migrate into peripheral tissues where they differentiate into mature mast cells.36 Interestingly, Irf8−/− mice had severely reduced numbers of bone marrow MCPs, but retained normal counts of intestinal MCPs (Figure 2; see “Discussion”).
IRF8 is expressed in GPs, but not in basophils, mast cells, and basophil/mast cell-restricted progenitor cells
Next the expression of IRF8 protein was examined on a per cell basis in basophils, mast cells, and their progenitor populations by utilizing IRF8-green fluorescent protein (GFP) chimera knock-in mice (Irf8Irf8Gfp/WT). The IRF8-GFP chimera protein is as functional as untagged IRF8,38 and the knock-in mice showed no abnormality in hematopoiesis.26,39 Somewhat unexpectedly, IRF8 expression, judged by GFP intensity, was not detectable in basophils in the bone marrow, spleen, and peripheral blood, or in mast cells in the peritoneal cavity, although it was abundant in B cells (Figure 3A). Neutrophils and eosinophils did not express IRF8 either, consistent with previous reports.40,41 In progenitor populations, a majority of GPs clearly express IRF8, but BaPs, pre-BMPs, MCPs, and BMCPs did not. In fact, Irf8 transcripts were expressed only in GPs (Figure 3B). These results, together with the finding that Irf8−/− mice retained GPs, but lost basophil/mast cell-restricted progenitor populations in the bone marrow, suggest that IRF8 may act in GPs to regulate the generation of downstream progenitors.
IRF8 protein expression in granulocytes, mast cells, and their progenitors. (A) Using Irf8Irf8Gfp/WT mice, IRF8 protein expression was determined as GFP fluorescence intensity in each indicated cell population gated by cell surface antigen staining using flow cytometry. Data are representative of 2 independent experiments with similar results. (B) Irf8 transcript levels were measured by RT-qPCR. Values in the bar graphs are means ± standard deviations from triplicate samples. Analysis of IRF8 protein expression in other hematopoietic progenitor populations has been reported elsewhere.26,27
IRF8 protein expression in granulocytes, mast cells, and their progenitors. (A) Using Irf8Irf8Gfp/WT mice, IRF8 protein expression was determined as GFP fluorescence intensity in each indicated cell population gated by cell surface antigen staining using flow cytometry. Data are representative of 2 independent experiments with similar results. (B) Irf8 transcript levels were measured by RT-qPCR. Values in the bar graphs are means ± standard deviations from triplicate samples. Analysis of IRF8 protein expression in other hematopoietic progenitor populations has been reported elsewhere.26,27
IRF8 is required for the development of basophils in a progenitor cell-intrinsic manner
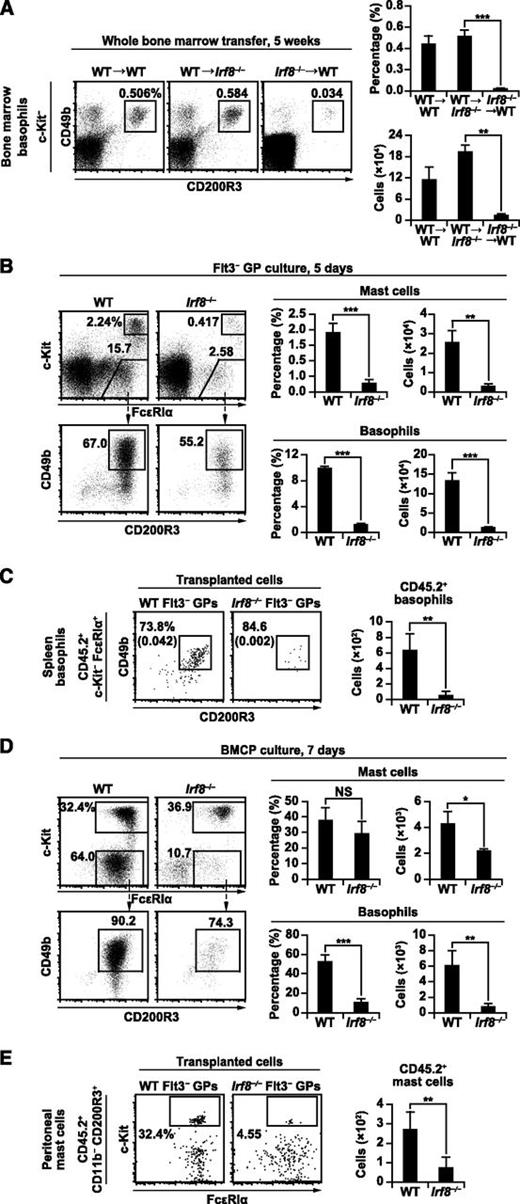
To examine whether the defect in basophil differentiation in Irf8−/− mice is intrinsic to bone marrow-derived cells, WT or Irf8−/− bone marrow cells were transplanted into irradiated Irf8−/− or WT recipients. Engraftment of WT bone marrow cells in Irf8−/− recipients restored the generation of basophils to normal levels. In contrast, engraftment of Irf8−/− bone marrow cells in WT mice failed to restore basophil generation (Figure 4A). In addition, we performed competitive mixed bone marrow chimera experiments. The percentages of basophils, eosinophils, pre-BMPs, and BaPs derived from Irf8−/− bone marrow cells were significantly reduced compared with those from WT cells (supplemental Figure 3). In contrast, the percentages of neutrophils derived from Irf8−/− bone marrow cells increased.
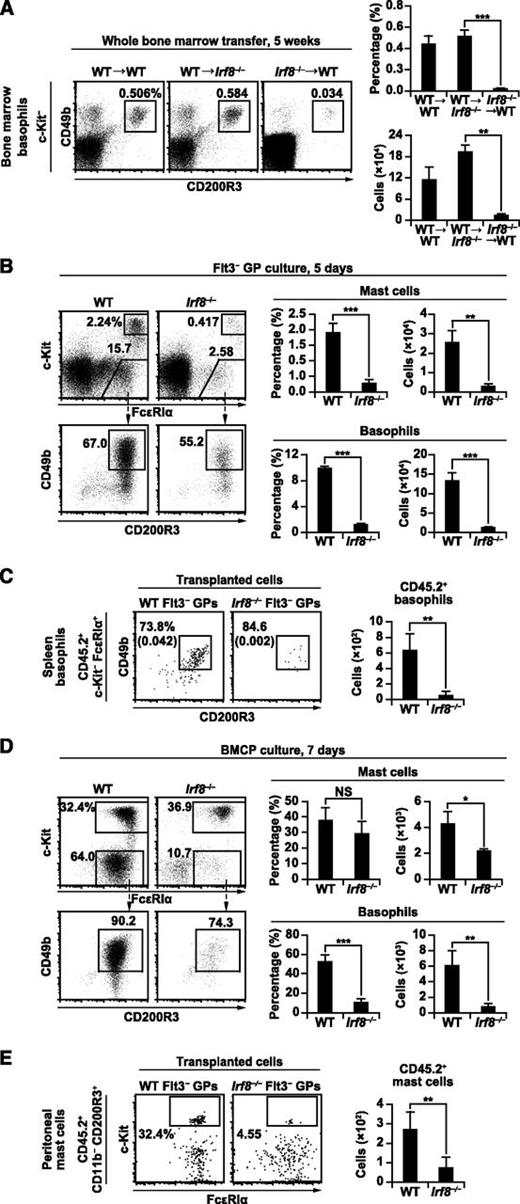
Bone marrow progenitor cell-intrinsic role of IRF8 in basophil and mast cell development. (A) Bone marrow transplantation experiments. Irradiated WT recipients reconstituted with WT bone marrow cells (WT→WT), irradiated Irf8−/− recipients reconstituted with WT bone marrow cells (WT→Irf8−/−), and irradiated WT recipients reconstituted with Irf8−/− bone marrow cells (Irf8−/−→WT) for 5 weeks were analyzed as in Figure 1A. Values in the bar graph are means ± standard deviations from 4 mice. Similar results were obtained in 3 other independent experiments. (B,D) fluorescence-activated cell sorter–purified Flt3– GPs (B) and splenic BMCPs (D) from WT and Irf8−/− mice were cultured in the presence of IL-3 for 5 or 7 days, respectively, and analyzed by flow cytometry. Data are representative of 3 independent experiments with similar results, and values in the bar graphs are means ± standard deviations from triplicate samples. (C,E) GP transfer experiments were performed to analyze the generation of basophils (C) and mast cells (E). For basophils (C), 10 000 Flt3− GPs from WT or Irf8−/− mice (CD45.2+) were transplanted into irradiated Ly5.1 mice (CD45.2−). Donor-derived (CD45.2+) splenic basophils were analyzed on day 4. Numbers in parentheses indicate the percentages relative to total cell counts. For mast cells (E), 20 000 Flt3− GPs (CD45.2+) with 3 × 105 Ly5.1 bone marrow cells (CD45.2−) were transplanted into irradiated mast cell-deficient KitW-sh/W-sh mice. CD45.2+ mast cells in the peritoneal cavity were analyzed 5 weeks after transplantation. Values in the bar graph are the mean ± standard deviation of 2 independent experiments with a total of 4 mice per group. *P < .05; **P < .01; ***P < .001 (Student t test). NS, not significant.
Bone marrow progenitor cell-intrinsic role of IRF8 in basophil and mast cell development. (A) Bone marrow transplantation experiments. Irradiated WT recipients reconstituted with WT bone marrow cells (WT→WT), irradiated Irf8−/− recipients reconstituted with WT bone marrow cells (WT→Irf8−/−), and irradiated WT recipients reconstituted with Irf8−/− bone marrow cells (Irf8−/−→WT) for 5 weeks were analyzed as in Figure 1A. Values in the bar graph are means ± standard deviations from 4 mice. Similar results were obtained in 3 other independent experiments. (B,D) fluorescence-activated cell sorter–purified Flt3– GPs (B) and splenic BMCPs (D) from WT and Irf8−/− mice were cultured in the presence of IL-3 for 5 or 7 days, respectively, and analyzed by flow cytometry. Data are representative of 3 independent experiments with similar results, and values in the bar graphs are means ± standard deviations from triplicate samples. (C,E) GP transfer experiments were performed to analyze the generation of basophils (C) and mast cells (E). For basophils (C), 10 000 Flt3− GPs from WT or Irf8−/− mice (CD45.2+) were transplanted into irradiated Ly5.1 mice (CD45.2−). Donor-derived (CD45.2+) splenic basophils were analyzed on day 4. Numbers in parentheses indicate the percentages relative to total cell counts. For mast cells (E), 20 000 Flt3− GPs (CD45.2+) with 3 × 105 Ly5.1 bone marrow cells (CD45.2−) were transplanted into irradiated mast cell-deficient KitW-sh/W-sh mice. CD45.2+ mast cells in the peritoneal cavity were analyzed 5 weeks after transplantation. Values in the bar graph are the mean ± standard deviation of 2 independent experiments with a total of 4 mice per group. *P < .05; **P < .01; ***P < .001 (Student t test). NS, not significant.
Then, GPs from WT and Irf8−/− mice were purified and cultured in the presence of IL-3, which is known to induce basophil and mast cell development in vitro.17 Our results indicate that Flt3− GPs generate basophils more efficiently than Flt3+ GPs, consistent with a previous report.15 The generation of basophils from both GP subpopulations on day 5 was severely impaired in the absence of IRF8 (Figure 4B; supplemental Figures 4A-B). To evaluate the differentiation potential of a single GP, Flt3− GPs from WT and Irf8−/− mice were single cell-sorted into 96-well plates and cultured in the presence of IL-3 for 5 days, and the resulting populations were analyzed by flow cytometry. As expected, basophils were efficiently generated from WT GPs, but that potential was severely impaired in Irf8−/− GPs (supplemental Figures 4C-D). Consistent with the known role of IRF8, Irf8−/− GPs produced more neutrophils (in the presence of IL-3) and less eosinophils (in the presence of IL-5 and stem cell factor) than WT GPs (supplemental Figures 4E-F). Transplantation of WT and Irf8−/− Flt3– GPs into irradiated mice further confirmed that Irf8−/− GPs do not produce basophils efficiently (Figure 4C). These results indicate that the defective basophil development seen in Irf8−/− mice is intrinsic to GPs in the bone marrow.
The role of IRF8 in the development of mast cells
Interestingly, Irf8−/− GPs produced fewer mast cells than WT GPs during the early culture phase (day 5), although mast cell counts in WT and Irf8−/− cultures reached comparable levels at 4 weeks (Figure 4B; supplemental Figures 4B and 5A). Irf8−/− Lin− cells also produced lower numbers of mast cells than WT Lin− cells during the early stage of in vitro cultures (see control [Ctrl]-transduced cells in Figures 5A and 7D).
The effect of forced IRF8 expression on basophil and mast cell differentiation. (A-B) Bone marrow Lin− cells and Flt3− GPs from WT and Irf8−/− mice were transduced with empty MIG (MSCV-internal ribosome entry site-GFP) or MIG-IRF8 retrovirus in the presence of IL-3 on day 1. Retrovirally transduced (GFP+) cells were analyzed for basophil and mast cell generation on day 7. (C) Bone marrow Lin− cells from Irf8−/− mice were transduced with empty MICD8 (MSCV-internal ribosome entry site-human truncated CD8) vector (Ctrl), MICD8-IRF8, MICD8-IRF8K79E, or MICD8-IRF8R289E, and analyzed as described in (A). Retrovirally transduced (human CD8+) cells were analyzed. Values in the bar graphs are means ± standard deviations from triplicate samples. Similar results were obtained in 3 additional independent experiments (A) or 1 additional independent experiment (B-C). *P < .05; **P < .01; ***P < .001 (Student t test).
The effect of forced IRF8 expression on basophil and mast cell differentiation. (A-B) Bone marrow Lin− cells and Flt3− GPs from WT and Irf8−/− mice were transduced with empty MIG (MSCV-internal ribosome entry site-GFP) or MIG-IRF8 retrovirus in the presence of IL-3 on day 1. Retrovirally transduced (GFP+) cells were analyzed for basophil and mast cell generation on day 7. (C) Bone marrow Lin− cells from Irf8−/− mice were transduced with empty MICD8 (MSCV-internal ribosome entry site-human truncated CD8) vector (Ctrl), MICD8-IRF8, MICD8-IRF8K79E, or MICD8-IRF8R289E, and analyzed as described in (A). Retrovirally transduced (human CD8+) cells were analyzed. Values in the bar graphs are means ± standard deviations from triplicate samples. Similar results were obtained in 3 additional independent experiments (A) or 1 additional independent experiment (B-C). *P < .05; **P < .01; ***P < .001 (Student t test).
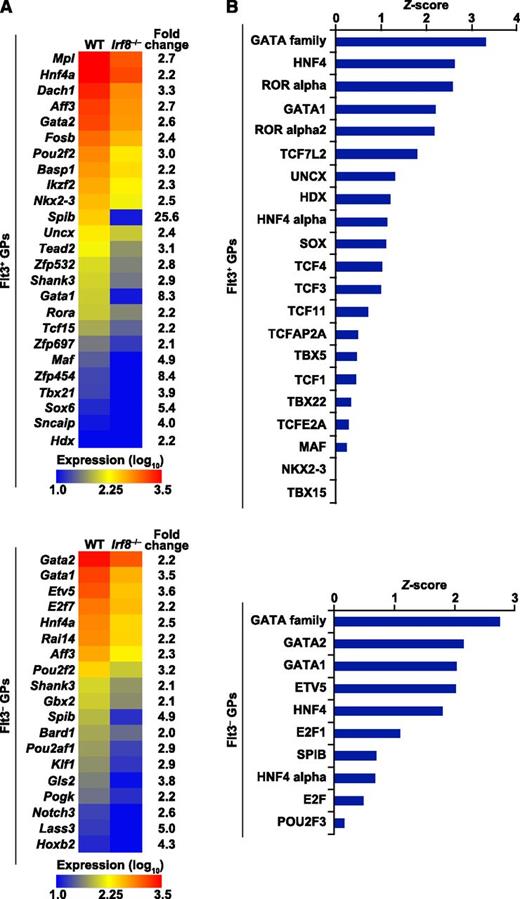
Transcriptome analysis of GPs and the prediction of transcription factors downstream of IRF8. Gene expression profiling of fluorescence-activated cell sorter–purified Flt3+ and Flt3− GPs from WT and Irf8−/− mice was performed by microarray in biological duplicates. (A) Expression levels of transcription factor genes downregulated in Irf8−/− GPs (false discovery rate <0.05; fold change >2) are displayed as a heat map. (B) Known DNA-binding motif analysis for the transcription factors depicted in (A) was performed in the 5 kb regions upstream of TSSs of the genes downregulated in Irf8−/− GPs (false discovery rate < 0.05; fold change > 2) in comparison with all the other genes. The significance of the enrichment of DNA-binding motif instances was quantified by the Z-score.
Transcriptome analysis of GPs and the prediction of transcription factors downstream of IRF8. Gene expression profiling of fluorescence-activated cell sorter–purified Flt3+ and Flt3− GPs from WT and Irf8−/− mice was performed by microarray in biological duplicates. (A) Expression levels of transcription factor genes downregulated in Irf8−/− GPs (false discovery rate <0.05; fold change >2) are displayed as a heat map. (B) Known DNA-binding motif analysis for the transcription factors depicted in (A) was performed in the 5 kb regions upstream of TSSs of the genes downregulated in Irf8−/− GPs (false discovery rate < 0.05; fold change > 2) in comparison with all the other genes. The significance of the enrichment of DNA-binding motif instances was quantified by the Z-score.
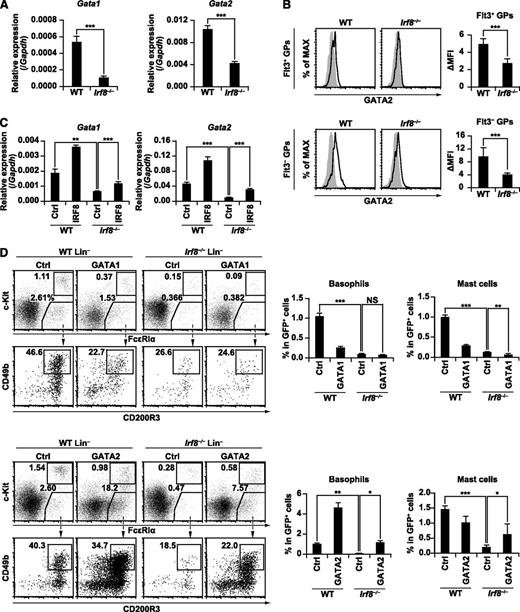
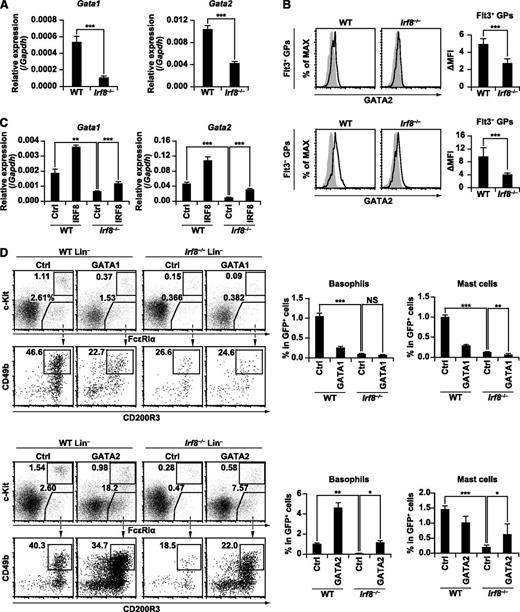
The requirement for IRF8 in GATA1 and GATA2 expression, and the effect of GATA1 and GATA2 transduction on basophil and mast cell development in Irf8−/− progenitor cells. (A) Gata1 and Gata2 mRNA expression in WT and Irf8−/− GPs was measured by RT-qPCR. GPs from each genotype were sorted into 3 separate tubes and analyzed by RT-qPCR. Values in the bar graph are means ± standard deviations. (B) GATA2 protein expression in WT and Irf8−/− GPs was examined by immunostaining followed by flow cytometry. ΔMFI (mean fluorescent intensity) was calculated by subtracting the background fluorescence intensity. Values in the bar graphs are means ± standard deviations from 3 independent experiments. (C) Gata1 and Gata2 mRNA expression in WT and Irf8−/− Lin− cells transduced with empty MICD8 vector (Ctrl) or MICD8-IRF8 as in Figure 5A. Transduced cells were purified by the magnetic-activated cell sorting (MACS) system on day 5 and analyzed by RT-qPCR. Values in the bar graphs are means ± standard deviations from triplicate samples. (D) Bone marrow Lin− cells from WT and Irf8−/− mice were transduced with empty MIG vector (Ctrl), MIG-GATA1, or MIG-GATA2 as in Figure 5A. Retrovirally transduced (GFP+) cells were analyzed for basophil and mast cell generation on day 5. Values in the bar graphs are means ± standard deviations from triplicate samples. Data are representative of 2 independent experiments with similar results. *P < .05; **P < .01; ***P < .001 (Student t test). NS, not significant.
The requirement for IRF8 in GATA1 and GATA2 expression, and the effect of GATA1 and GATA2 transduction on basophil and mast cell development in Irf8−/− progenitor cells. (A) Gata1 and Gata2 mRNA expression in WT and Irf8−/− GPs was measured by RT-qPCR. GPs from each genotype were sorted into 3 separate tubes and analyzed by RT-qPCR. Values in the bar graph are means ± standard deviations. (B) GATA2 protein expression in WT and Irf8−/− GPs was examined by immunostaining followed by flow cytometry. ΔMFI (mean fluorescent intensity) was calculated by subtracting the background fluorescence intensity. Values in the bar graphs are means ± standard deviations from 3 independent experiments. (C) Gata1 and Gata2 mRNA expression in WT and Irf8−/− Lin− cells transduced with empty MICD8 vector (Ctrl) or MICD8-IRF8 as in Figure 5A. Transduced cells were purified by the magnetic-activated cell sorting (MACS) system on day 5 and analyzed by RT-qPCR. Values in the bar graphs are means ± standard deviations from triplicate samples. (D) Bone marrow Lin− cells from WT and Irf8−/− mice were transduced with empty MIG vector (Ctrl), MIG-GATA1, or MIG-GATA2 as in Figure 5A. Retrovirally transduced (GFP+) cells were analyzed for basophil and mast cell generation on day 5. Values in the bar graphs are means ± standard deviations from triplicate samples. Data are representative of 2 independent experiments with similar results. *P < .05; **P < .01; ***P < .001 (Student t test). NS, not significant.
Our findings that Irf8−/− mice lack pre-BMPs and MCPs in the bone marrow, but retain splenic BMCPs and peripheral tissue MCPs, and mast cells led us to speculate that MCPs and mast cells in peripheral tissues of Irf8−/− mice might be derived from BMCPs. To test this hypothesis, we purified BMCPs from the spleens of WT or Irf8−/− mice and cultured them with IL-3. Unexpectedly, Irf8−/− BMCPs proliferated poorly and generated a reduced number of basophils and, to a lesser degree, mast cells compared with WT BMCPs (Figure 4D; supplemental Figure 5B). The process by which IRF8 affects the developmental potential of BMCPs, which do not themselves express IRF8, is unknown; one theory would be that the IRF8 deficiency causes epigenetic changes at an earlier stage of differentiation.
Overall, our results suggest that Irf8−/− progenitors are not fully capable of generating mast cells in vitro, which differs somewhat from the in vivo data demonstrating normal mast cell counts in tissues. Mast cells are long-lived and known to proliferate in peripheral tissues.42 Therefore, we were intrigued by the effect of nonmyeloid Irf8−/− cells. Previous studies have demonstrated that the serum IgE levels in Irf8−/− mice are approximately 70 times higher than those in WT mice,43 and IgE is known to enhance mast cell survival.44 This prompted us to examine whether Irf8−/− GPs themselves efficiently produce mast cells in vivo. To accomplish this, purified Flt3− GPs from WT and Irf8−/− mice were transplanted into irradiated KitW-sh/W-sh mice deficient in mast cells, and the recipients were analyzed 5 weeks later. Our results indicate that Irf8−/− GPs failed to efficiently generate mast cells in vivo when the other hematopoietic cell lineages were of Irf8+/+ origin (Figure 4E). Overall, these results suggest that IRF8 does play a role in promoting the development of mast cells.
Introducing IRF8 into Irf8−/− GPs restores the production of basophils and mast cells
To examine if the restoration of IRF8 expression could rescue basophil and mast cell differentiation from Irf8−/− progenitor cells in the short-term culture, WT and Irf8−/− Lin− cells or GPs from the bone marrow were inoculated in the presence of IL-3, retrovirally transduced with empty MIG (murine stem cell virus [MSCV]-internal ribosome entry site-GFP) vector or MIG-IRF8, and further cultured until day 7 (Figures 5A-B). As expected, the resulting percentages of basophils and mast cells were significantly lower in empty MIG-transduced Irf8−/− culture than in empty MIG-transduced WT culture. However, IRF8 transduction into Irf8−/− culture resulted in a significant increase in the percentages of basophils and mast cells, reaching levels comparable to those in empty MIG-transduced WT culture, whereas the differentiation of neutrophils was inhibited (supplemental Figure 6). Additionally, IRF8 transduction into WT cells further boosted the generation of basophils on day 7. Importantly, the introduction of mutant IRF8 (ie, K79E [a point mutant in the DNA binding domain] and R289E [a point mutant in the protein-protein interaction module]) into Irf8−/− Lin− cells failed to rescue basophil and mast cell differentiation (Figure 5C). The expression of transduced IRF8 was detected by intracellular immunostaining and flow cytometry (supplemental Figure 7). These results further confirmed the progenitor cell-intrinsic role of IRF8.
To examine whether basophils rescued by IRF8 transduction are functional, we stimulated basophils derived from IRF8-transduced WT or Irf8−/− Lin− cells with phorbol 12-myristate 13-acetate and ionomycin, and we performed intracellular IL-4 staining. IL-4 was clearly produced in the c-Kit− CD49b+ FcεRIα+ basophil population, regardless of their genotype and IRF8 transduction (supplemental Figure 8).
Prediction of transcription factors that act downstream of IRF8
The above findings suggest that IRF8 acts in GPs, presumably by inducing its downstream genes. We speculated that such genes would include those encoding transcription factors, which continue to be expressed during basophil and mast cell differentiation. Therefore, transcriptome microarray analysis was performed in purified Flt3+ and Flt3− GPs from WT and Irf8−/− mice (see supplemental Figure 9 for correlation matrix and scatter plots). Multiple transcription factor genes were downregulated in Irf8−/− GPs, suggesting that these genes depend on IRF8 for normal expression (Figure 6A). To predict which transcription factors among them were likely to act downstream of IRF8 and critically regulate basophil and mast cell development, the enrichment of their binding motifs in the promoter regions (5 kb upstream of the transcription start sites [TSSs]) of the genes downregulated in Irf8−/− GPs was calculated in comparison with that in the promoter regions of other genes. The binding motifs of GATA transcription factors were most significantly enriched, displaying the highest Z-score both in Flt3+ and Flt3− GPs (Figure 6B). Interestingly, both GATA1 and GATA2 have been shown to promote basophil and mast cell development.20-22 These results suggest that GATA transcription factors may mediate the action of IRF8 in basophil/mast cell development.
IRF8 upregulates the expression of Gata1 and Gata2
Consistent with the microarray data, RT-qPCR confirmed that the expression of Gata1 and Gata2 transcripts was significantly lower in Irf8−/− GPs (Figure 7A). Intracellular immunostaining and flow cytometry demonstrated that GATA2 protein expression was lower in both Flt3+ and Flt3− GPs from Irf8−/− mice when compared with those from WT mice (Figure 7B). However, GATA1 protein expression was not evident in GPs, although readily detectable in megakaryocyte-erythroid progenitors (supplemental Figure 10). Together with the RT-qPCR data in Figure 7A, this data suggests that GATA1 expression is relatively low in GPs. The expression of GATA1 and GATA2 in IRF8-transduced Lin− cells in the presence of IL-3 on day 5 was then measured. GATA1 and GATA2 expression in control vector-transduced Irf8−/− culture was lower than that in control vector-transduced WT culture at both the transcriptional and protein levels, but this defect was rescued with the transduction of IRF8 (Figure 7C; supplemental Figure 11). Additionally, IRF8 expression augmented GATA1 and GATA2 expression in WT cells. These findings suggest that IRF8 induces Gata1 and Gata2 genes directly or indirectly and is indispensable for their normal expression in GPs.
Forced expression of GATA2, but not GATA1, in Irf8−/− progenitor cells rescues the development of basophils and mast cells
To test if GATA1 and/or GATA2 mediate the functions of IRF8 in basophil and mast cell development, GATA1 or GATA2 was transduced into Irf8−/− bone marrow Lin− cells in the presence of IL-3 and analyzed on day 5. The expression of transduced GATA1 and GATA2 was confirmed by intracellular immunostaining and flow cytometry (supplemental Figure 12). We found that GATA2, but not GATA1, rescued the differentiation of basophils and, at least partially, also the early generation of mast cells (Figure 7D; supplemental Figure 13A). The percentages of neutrophils were reduced by both GATA1 and GATA2 transduction in Irf8−/− cells (supplemental Figure 13B).
Of note, GATA2 also promoted the generation of basophils in WT Lin− cells. Thus, one may argue that the forced expression of GATA2 acts on a pathway independent of IRF8. For example, overexpressed GATA2 might have simply promoted the proliferation of basophils or their progenitors. However, the analysis of carboxyfluorescein diacetate succinimidyl ester-stained Lin− cells demonstrated that GATA2-transducion did not augment carboxyfluorescein diacetate succinimidyl ester dilution in basophils, suggesting that GATA2 stimulated the differentiation toward, rather than the expansion of, basophil lineage cells (supplemental Figure 13C). The fact that IRF8 also enhanced the generation of basophils, concomitant with the upregulated expression of GATA2 in WT Lin− cells (above; Figures 5A and 7C), further suggests the tight relationship between IRF8 and GATA2. Overall, our data suggest that GATA2 mediates the activity of IRF8 in the development of basophils and, at least partially, mast cells.
Discussion
Implications for the developmental pathways of basophils and mast cells
In this study, we demonstrated that Irf8−/− mice lack basophils, but retain mast cells in vivo. We also showed that IRF8 is expressed in GPs, but not in basophils, mast cells, and basophil/mast cell-restricted progenitors.
The defect in basophil development in Irf8−/− mice may be due to the absence of pre-BMPs and by the inability of Irf8−/− GPs and BMCPs to generate basophils. The role of IRF8 in mast cell development is somewhat complicated. Irf8−/− mice exhibited a severe reduction in bone marrow pre-BMPs and MCPs, while retaining splenic BMCP, peripheral tissue MCPs, and mast cells. However, Lin− cells, GPs, and BMCPs from Irf8−/− mice were less efficient than their WT counterparts in generating both basophils and mast cells in vitro. Moreover, transplanted Irf8−/− GPs produced a reduced number of both basophils and mast cells compared with WT GPs in vivo. These results suggest that IRF8 also contributes to the development of mast cells.
Then, how do Irf8−/− peripheral tissue MCPs and mast cells develop? Because mast cells have been shown to proliferate in peripheral tissues and are long-lived,42 a small number of MCPs and/or mast cells, initially derived from Irf8−/− BMCPs or residual pre-BMPs, may proliferate and eventually reach normal levels. This appeared to be replicated in the long-term in vitro mast cell culture experiments supplemented with the potent cytokine IL-3. In addition, the extremely high IgE levels in Irf8−/− mice may help enhance the survival of mast cells and peripheral tissue MCPs, both of which express FcεRIα.17,36,44
Based on our current data and the recognized developmental potential of the progenitor populations, it is tempting to speculate that pre-BMPs are derived from GPs, and bone marrow MCPs are derived from pre-BMPs. However, such theories regarding the relationships between progenitor populations require further and extensive study.
Interestingly, the complete blood counts from a human patient with immunodeficiency caused by the K108E mutation of IRF8 failed to detect peripheral blood basophils,45 suggesting a conserved role for IRF8 in mice and humans.
Coordinated regulation of basophil and mast cell development by multiple transcription factors
Here we discuss the possible relationships between IRF8 and some of other transcription factors known to be critical for basophil and mast cell development, namely GATA2, STAT5, C/EBPα, and MITF. GATA2 expression has been reported to be higher in BaPs, MCPs, basophils, and mast cells than in GMPs,16,22 and our current study has identified IRF8 as a factor mediating this elevation by acting in GPs. However, IRF8 expression is absent in pre-BMPs, BaPs, and MCPs. Therefore, Gata2 expression is likely maintained through an IRF8-independent mechanism, possibly by GATA2 itself.46 Interestingly, both IRF8 and GATA2 inhibited the in vitro development of neutrophils from Irf8−/− bone marrow Lin− cells, while promoting that of basophils and mast cells. Thus, the IRF8-GATA2 axis may couple the reciprocal regulation of the development of neutrophil and basophil/mast cell lineages from GPs. It is possible that STAT5 is responsible for the repression of Irf8 expression in pre-BMPs. STAT5 has been shown to induce Cebpa or Mitf in pre-BMPs and direct them to differentiate into basophils and mast cells, respectively; in DC progenitors, STAT5 represses Irf8 to inhibit plasmacytoid DC development.16,47 Based on these data, we propose the following model for basophil and mast cell development: IRF8 induces Gata2 in GPs; once induced, Gata2 upregulates or maintains its expression via self-activation; in pre-BMPs, STAT5 is activated to repress Irf8, while inducing Cebpa or Mitf. Recently, we reported that IRF8 inhibits the transcriptional activity of C/EBPα in mononuclear phagocyte progenitors to prevent neutrophil differentiation.27 Because the expression level of IRF8 is much lower in GPs than in mononuclear phagocyte progenitors, we infer that the inhibition of C/EBPα by IRF8 in GPs (and retrovirally transduced IRF8) is limited enough to permit them to differentiate into basophils. In addition, we discuss the subject of basophila in chronic myelogenous leukemia in relation to IRF8, the requirement for IRF8 in Gata1 expression in GPs, and the possible role of IRF8 in regulating the development of multiple antigen presenting cell types (supplemental Discussion).
The IRF8R289E mutant failed to restore the development of basophils and mast cells, suggesting that interaction with another transcription factor(s) was required. IRF8 has been shown to interact with several transcription factors including PU.1, IRF1, IRF2, and basic leucine zipper transcription factor, ATF-like (BATF) on DNA.24,48 PU.1, a critical partner in monocyte/DC differentiation, is a good candidate because PU.1-deficient embryos and neonates lack granulocytes.49,50 Because Irf2−/− mice exhibit basophil expansion,6 the possibility of an antagonistic relationship between IRF8 and IRF2 is also intriguing.
Induction of Gata2 by IRF8
Our results clearly indicated the requirement for IRF8 in Gata2 expression in GPs. However, the detailed mechanism by which IRF8 induces Gata2 remains unknown. Because the induction of Gata2 expression and basophil differentiation by IRF8 is relatively slow (Figure 7C), we hypothesize that IRF8 may induce an intermediate factor that directly binds to the Gata2 promoter or enhancer, resulting in the induction of Gata2 during basophil/mast cell differentiation. We initially attempted to identify cell lines suitable for reporter assays or chromatin immunoprecipitation–sequencing (ChIP-seq), however all the cell lines tested failed to induce Gata2 upon the introduction of IRF8. It is possible that the factor connecting IRF8 and Gata2 is inducible only in cells possessing basophil/mast cell differentiation potential. Future ChIP-seq analysis to examine histone modifications, using freshly isolated WT and Irf8−/− GPs would identify IRF8-dependent enhancer elements to which “the connecting factor” binds. This factor may be one of the transcription factors displaying decreased gene expression in Irf8−/− GPs (Figure 6A). Our observation that the residual basophils generated from Irf8−/− progenitor cells expressed GATA2 (supplemental Figure 12) also supports the notion that IRF8 may not be the direct transactivator of Gata2.
We note here that we have performed a preliminary experiment for one candidate transcription factor, SpiB. The expression of Spib was downregulated in Irf8−/− GPs at the highest fold-change in both Flt3+ and Flt3− GPs. When we tested Spib−/− mice, however, the numbers of basophils were comparable to those in WT mice, suggesting that SpiB is unlikely to be a major regulator of basophil development (supplemental Figure 14). This result also emphasizes the validity of the in silico prediction of candidate transcription factors by motif analysis (Figure 6B), which showed a relatively low Z-score for SpiB.
In conclusion, our study has unveiled an indispensable and previously unrecognized role of IRF8 in the development of basophils and mast cells. These results further elucidate the mechanism involved in generating these critical myeloid populations, and contribute to a comprehensive understanding of the role of IRF8 in myeloid development.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Jun Nakabayashi, Dr Masatoshi Nakazawa, Masahiro Yoshinari, and Naofumi Kaneko at Yokohama City University for their help with experiments; Dr Kazuyoshi Takeda at Juntendo University for valuable suggestions; and Dr Susumu Nakae at the University of Tokyo for providing KitW-sh/W-sh mice.
This work was supported by KAKENHI grants-in-aid (24390246 and 24118002) (T.T.) and (24790322) (D.K.) from the Japan Society for the Promotion of Science, the fund for Creation of Innovation Centers for Advanced Interdisciplinary Research Areas Program in the Project for Developing Innovation Systems from MEXT (T.T.), a grant for Strategic Research Promotion from Yokohama City University (T.T.), and the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (H.W., H.C.M.) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (K.O.), and the supercomputing resource was provided by Human Genome Center of the Institute of Medical Science at the University of Tokyo.
Authorship
Contribution: H.Sas., D.K., and T.T. designed research; H. Sas., D.K., I.S., S.-i.K., C.K., H.Sat., and A.N. performed experiments; H.Sas., D.K., N.O., A.N., H.A., and T.T. analyzed data; H.Sas., D.K., and T.T. wrote the manuscript; H.W., T.K., H.C.M., and K.O. provided critical materials; and T.T. supervised the project.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tomohiko Tamura, Department of Immunology, Yokohama City University Graduate School of Medicine, 3-9 Fukuura, Kanazawa-ku, Yokohama 236-0004, Japan; e-mail: tamurat@yokohama-cu.ac.jp.
References
Author notes
H.S. and D.K. contributed equally to this work.