In this issue of Blood, de Vries et al report the contribution of genetic variants to plasma ADAMTS13 (disintegrin and metalloproteinase with thrombospondin motifs 13) activity with a hypothesis-free genome-wide association (GWA) approach using the Rotterdam Study, a large population-based cohort study.1
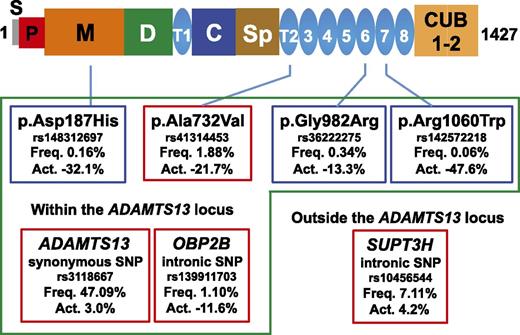
ADAMTS13 domain structure and functional genetic variants. Variants boxed with green are located within the ADAMTS13 locus. Variants boxed with red are significantly associated common variants among genome-wide SNPs. Variants boxed with blue are significantly associated rare variants among exome-wide SNPs. Act., percentage of increased or decreased plasma ADAMTS13 activity; C, Cys-rich domain; CUB, complement C1r/C1s; D, disintegrin-like domain; Freq., allele frequency; M, metalloprotease domain; P, propeptide; S, signal peptide sequence; Sp, spacer domain; T, thrombospondin type 1 repeat.
ADAMTS13 domain structure and functional genetic variants. Variants boxed with green are located within the ADAMTS13 locus. Variants boxed with red are significantly associated common variants among genome-wide SNPs. Variants boxed with blue are significantly associated rare variants among exome-wide SNPs. Act., percentage of increased or decreased plasma ADAMTS13 activity; C, Cys-rich domain; CUB, complement C1r/C1s; D, disintegrin-like domain; Freq., allele frequency; M, metalloprotease domain; P, propeptide; S, signal peptide sequence; Sp, spacer domain; T, thrombospondin type 1 repeat.
ADAMTS13 is a plasma metalloprotease that specifically cleaves the plasma adhesive protein von Willebrand factor (VWF). VWF is a large plasma glycoprotein and mediates platelet adhesion at sites of vascular injury. VWF is synthesized in an ultralarge multimeric form that has a very strong platelet aggregation activity. VWF-induced platelet aggregation depends on its multimeric size, which is controlled by ADAMTS13.2 ADAMTS13 has a discrete domain structure, and the gene consists of 29 exons located on chromosome 9. Severe deficiency of ADAMTS13 activity due to ADAMTS13 variants or acquired autoantibodies that inhibit ADAMTS13 activity leads to thrombotic thrombocytopenic purpura (TTP), a disease caused by platelet aggregation by ultralarge VWF multimers. Rare causative loss-of-function genetic variants have been identified in patients with congenital TTP.3 In addition to TTP, ADAMTS13 may contribute to other thrombotic disorders. Low plasma levels of ADAMTS13 are associated with an increased risk for myocardial infarction and stroke. In addition, low plasma level is observed in severe sepsis, disseminated intravascular coagulation, and complicated malarial infection.4 These findings suggest a contribution of VWF-dependent platelet aggregation in some thrombotic disease states. If so, the identification of genetic and acquired factors that affect plasma ADAMTS13 activity is very important.
So far, rare variants of the ADAMTS13 gene causing TTP have been identified. In addition, a few common genetic variants with modest effects on ADAMTS13 are reported. However, it is not clear whether the genetic variants exhibit strong associations at the locus. Furthermore, genetic variations outside the ADAMTS13 locus remain unknown.
In this issue, de Vries et al adopted a systematic hypothesis-free GWA study approach to identify genetic variants that affect plasma ADAMTS13 activity by using a large, prospective, population-based cohort study, the Rotterdam Study.1 ADAMTS13 activity was measured using a fluorogenic peptidyl substrate (fluorescence resonance energy transfer substrate [FRETS]-VWF73)5 in >6000 individuals. Genome-wide single nucleotide polymorphisms (SNPs) for common genetic variants and exome-wide SNPs for rare genetic variants were genotyped. After careful examination of these genotyped data, de Vries et al identified p.Ala732Val (rs41314453) in ADAMTS13 as the strongest genetic determinant of ADAMTS13 activity; the minor allele was associated with a decrease of >20% (see figure). Furthermore, they identified independent associations with a common variant in SUPT3H outside the ADAMTS13 locus and 5 genetic variants at the ADAMTS13 locus. The variant p.Ala732Val in ADAMTS13 explained 3.6% to 6.5% of the variance in ADAMTS13 activity, which was comparable to the variance explained by age (3.9%-6.5%) or by sex (4.5%-6.7%). The 4 independently significant common SNPs (boxed with red in the figure) explained 5.8% to 8.2% of the variance.
The genetic variants influencing plasma ADAMTS13 activity have been mostly restricted to the ADAMTS13 locus (see figure). The only exception was SUPT3H outside the ADAMTS13 locus, and its effect was relatively small. This restriction is in sharp contrast to genetic factors influencing VWF. Genome-wide analysis for plasma VWF levels showed that multiple loci are involved.6 Various steps, including VWF synthesis, packaging into Weibel-Palade bodies, secretion, and removal from the circulation, are involved in determining VWF levels,7 and genetic variants of proteins involved in these steps could affect plasma VWF levels. ADAMTS13 activity may be regulated in a much simpler manner than VWF.
Because rare genetic variants have generally emerged relatively recently, they show greater geographic clustering than common variants. A previous study done in the Japanese population showed that p.Pro475Ser in the ADAMTS13 gene, restricted to the East Asian population and having an allele frequency of 5%, was associated with 14% decreased activity.8 In Northern and Central European countries, the E1382Rfs*6 mutation due to the 4143insA mutation is frequent among patients with TTP.9 The present study did not identify these variants, probably because all the participants were from a small geographic area.
As for the relation to congenital TTP or other thrombotic disorders, 2 rare genetic variants, p.Asp187His and p.Arg1060Trp, have previously been reported in patients with TTP. The present study revealed that their allele frequencies are 0.16% and 0.06%, respectively, indicating that a substantial number of individuals carry these variants in a homozygous or compound heterozygous state and have a genetic risk for TTP. Furthermore, it may be important to prospectively follow these variant heterozygous carriers to determine an increased risk for myocardial infarction or stroke. Thus, the results obtained from the GWA study for ADAMTS13 activity have a big impact for not only TTP but also other thrombotic disorders.
Conflict-of-interest disclosure: T.M. is employed by the National Cerebral and Cardiovascular Center, which has an awarded patent on the use of the reagent FRETS-VWF73.