Key Points
DCs and T-lineage cells in the thymus have separate origins.
Availability of microenvironmental niches in the thymus determines lineage fate.
Abstract
The origins of dendritic cells (DCs) and other myeloid cells in the thymus have remained controversial. In this study, we assessed developmental relationships between thymic dendritic cells and thymocytes, employing retrovirus-based cellular barcoding and reporter mice, as well as intrathymic transfers coupled with DC depletion. We demonstrated that a subset of early T-lineage progenitors expressed CX3CR1, a bona fide marker for DC progenitors. However, intrathymic transfers into nonmanipulated mice, as well as retroviral barcoding, indicated that thymic dendritic cells and thymocytes were largely of distinct developmental origin. In contrast, intrathymic transfers after in vivo depletion of DCs resulted in intrathymic development of non–T-lineage cells. In conclusion, our data support a model in which the adoption of T-lineage fate by noncommitted progenitors at steady state is enforced by signals from the thymic microenvironment unless niches promoting alternative lineage fates become available.
Introduction
T-cell development in the thymus is tightly regulated through the interactions of thymocytes with nonthymocytes, such as thymic epithelial cells and thymic dendritic cells (tDCs). tDCs consist of plasmacytoid DCs (pDCs) and two distinct populations of conventional DCs (cDCs) that can be distinguished based on differential expression of CD8α and Sirpα.1 In secondary lymphoid organs, both CD8α+ and CD8α− cDCs are derived from a common developmental pathway. This pathway is comprised of various progressively lineage restricted intermediates ranging from myeloid progenitors (MPs) via macrophage/DC progenitors (MDPs) and common DC progenitors (CDPs) to pre-DCs.2
However, the developmental origin of tDCs within the thymus has remained controversial. Thus, the discovery of DHJH immunoglobulin rearrangements in tDCs, but not in splenic CD8α+ DCs supported a lymphoid origin of tDCs.3 In addition, both human and mouse tDCs have been reported to express the pre-TCRα chain.4,5 Furthermore, early T-lineage progenitors (ETPs), which constitute the earliest detectable intrathymic T-cell precursors, have been shown to retain DC potential and CD8α+ tDCs develop intrathymically with kinetics paralleling those of T cells.6-10 However, analysis of mouse models expressing lineage-specific reporter genes or Cre recombinase to allow genetic fate mapping in vivo provided evidence for separate origins of thymocytes and tDCs. Thus, using Il7r-based fate mapping, which labels virtually all thymocytes, Schlenner et al showed that among tDCs, the majority of pDCs but only minor frequencies of other tDC subsets, were derived from progenitors with a history of Il7r expression.11 Consistently, fate mapping based on Ptcra expression showed that tDCs were not derived from T-lineage committed cells.12 An intrathymic DC progenitor population that is likely to be of independent origin from ETPs was identified in mice expressing a CD207(Langerin)-green fluorescent protein (GFP) reporter gene.13 In addition, these progenitors already expressed CD11c and low levels of MHC-II, and appeared therefore to be more mature than peripheral pre-DCs of the canonical DC developmental pathway.
To date, it remains difficult to reconcile these divergent findings. Whereas some experimental approaches, such as analysis of lineage potential ex vivo have directly been contested,14 some experiments supporting either one model or the other remain compelling.15 Lineage fate mapping, in combination with simultaneous deletion of Notch1 in mice indicated that in vivo, in the absence of Notch signaling, ETPs can be diverted to become tDCs.16 Absence of Notch1 is likely to lift Hes1-dependent repression of the key myeloid transcription factor gene, Cebpa.17 These experiments support the notion that ETPs are basically equipped with DC potential, but do not adopt DC fate in a microenvironment rich in Notch ligands, such as the thymus. However, this hypothesis remains to be tested with ETPs that retain an intrinsic capacity to develop into T cells.
In this study, we have assessed whether a pro-T-cell–derived DC progenitor exists in the thymus that can be diverted into the DC lineage in vivo. We identified and characterized a subset of ETPs expressing CX3CR1. Retroviral barcoding in combination with next-generation sequencing suggested that at steady state tDC and T-cell development are largely independent. Finally, we demonstrated that various progenitors, including ETPs and MP/MDPs can give rise to tDCs in vivo upon depletion of endogenous tDCs, but not in their presence. Taken together, our data support a model in which the thymic microenvironment provides a strong inductive force for T-cell development, thereby preventing diversion into alternative lineages. However, liberation of bona fide tDC niches generates a permissive state for intrathymic DC development from various progenitors, including pro-T cells.
Methods
Mice
C57BL/6J mice (CD45.2) and B6.SJL-PtprcaPepcb/BoyJ mice (termed “B6 CD45.1” throughout this study) were purchased from Charles River. CD11c.DOG mice expressing diphtheria toxin receptor under the control of the CD11c promoter were described previously18 and crossed to B6 CD45.1 to become CD45.1/CD45.2 heterozygous. All experiments were performed with heterozygous mice for the DOG construct. B6.129P-CX3CR1tm1Litt/J (CX3CR1GFP/+) reporter mice19 and C57BL/6J x B6 CD45.1 F1 mice (CD45.1/CD45.2 heterozygous) were bred at the animal facility of the Hannover Medical School. Animals were maintained under specific pathogen-free conditions. All animal experiments were conducted in accordance with local and institutional guidelines.
Antibodies and flow cytometry
Monoclonal antibodies specific for CD4 (RM4-5, GK1.5), CD8 (53-6.7), CD25 (PC61), CD44 (IM7), Gr-1 (RB6-8C5), erythroid cell marker (Ter-119), CD19 (1D3), CD11b (M1/70), pan-NK (DX5), CD45.1 (A20), CD45.2 (104), B220 (RA3-6B2), CD117 (ACK2), Sca-1 (E13-161.7), CD135 (A2F10), CD127 (A7R34), Ki-67 (B56), Sirpα (P84), and CD115 (AFS98) were used purified or as various fluorescent or biotin conjugates. Antibodies were purified from hybridoma supernatants or were purchased from eBioscience, BD Biosciences, or BioLegend. Phycoerythrin-Cy7 conjugated streptavidin (BD Biosciences) was used to reveal staining with biotinylated monoclonal antibody. Data were analyzed with FloJo software (Tree Star). Lin− cells were isolated from total bone marrow (BM) by staining cell suspensions with a lineage-specific antibody cocktail (anti-CD4, anti-CD8, anti-CD19, anti-CD11b, anti-Gr-1, Ter-119, and DX5), followed by incubation with anti–rat-IgG-conjugated magnetic beads (Dynal, Invitrogen) and magnetic bead depletion of mature lineages. Double-negative (DN) thymocytes were enriched by complement lysis of double-positive (DP) and single-positive (SP) cells using anti-CD4 and anti-CD8 antibodies (clones RL1.72 and 31M), followed by incubation with Low-Tox-M rabbit complement (Cedarlane). Complement lysis based on this protocol did not affect cells expressing low levels of CD4.
Cell lines
OP9 BM stromal cells expressing the Notch ligand Delta-like ligand 1 (OP9-DL1) and OP9-control cells (OP9-GFP) were provided by J.C. Zúñiga-Pflücker (University of Toronto, Toronto, Canada).20 BaF3 cells were cultured in RPMI medium supplemented with 10% fetal calf serum, 100 U/mL penicillin, 0.1 mg/mL streptomycin, and IL-3 (10 ng/mL).
OP9 cocultures
OP9 coculture assays were essentially performed as described.20 Precursors were plated at an initial density of 1 to 5 × 102 cells onto subconfluent OP9-GFP or OP9-DL1 monolayers at 5 × 104 cells/well in a 24-well plate. All cocultures were performed in the presence of 1 ng/mL IL-7, 5 ng/mL Flt3 ligand (Flt3-L), and 10 ng/mL stem cell factor (SCF) for OP9-DL1 assays and 0.1 ng/mL IL-7, 100 ng/mL Flt3-L, 10 ng/mL SCF and 5% of granulocyte macrophage (GM)–colony-stimulating factor (CSF) supernatant prepared in house (corresponding to 12-22 ng/mL as assessed by enzyme-linked immunosorbent assay) for OP9-GFP cocultures. Fifty percent of medium was exchanged at days 4 and 7 of coculture.
Intrathymic transfers
Two to 10 × 103 CX3CR1− ETPs, CX3CR1+ ETPs, or MP/MDPs isolated from CX3CR1GFP/+ mice were injected into thymi of nonirradiated B6 CD45.1/CD45.2 or CD11c.DOG mice 1 day after diphtheria toxin (DT) (8 μg/gbw) treatment. Thymi were analyzed for donor-derived cells 21 days after transfer.
Barcoded vector library
Design of the barcode library was performed analogous to the method described by Verovskaya et al.21 Briefly, bar code oligonucleotides of the sequence: GTACAAGTAANNATCNNGATSSAAANNGGTNNAACNNTGTAAAACGACGGCCAGTGAC (see supplemental Figure 1A, available on the Blood Web site) were inserted into the 3′UTR of the coding sequence for eGFP in the gammaretroviral SF91 vector.22,23 Approximately 6000 clones were collected. Re-transformation followed by Sanger sequencing of 86 clones revealed one duplicate and near-equal distribution of all variable bases (supplemental Figure 1B), indicating a depth of the library of at least 4000 barcodes.
Transduction of cells
For production of retroviral vectors, HEK293T cells were transfected with pCL-Eco (coexpressing gag, pol, and env derived from murine leukemia virus) and library-containing SF91 plasmids. Eight hours after transfection, medium was exchanged and supernatant containing retroviral particles was collected after 24, 48, and 72 hours posttransfection. Retroviral transductions of BaF3 cells were performed on 24-well plates (Sarstedt, Germany) in the presence of 8 μg/mL of polybrene (Sigma-Aldrich). Lin−Sca-1+CD117hi (LSK) cells were sorted from lineage-depleted BM and cultured overnight in α-minimum essential medium supplemented with SCF (50 ng/mL), IL-7 (25 ng/mL), Flt-3L (25 ng/mL), and IL-6 (20 ng/mL). LSK cells were then transferred into 96-well plates (Sarstedt, Germany) preloaded with retroviral vector attached to RetroNectin (Takara, Japan) according to the manufacturer’s protocol.
BM chimeras and cell analysis
Some 48 hours after transduction with library vector, GFPlo LSKs were sorted and a minimum of 50 000 LSK cells were IV transferred into lethally irradiated (9 Gy) C57BL/6 recipients. BM, spleen, and thymic cells were isolated 8 to 10 weeks after transfer. tDCs (CD11c+MHC-II+) were sorted after CD11c-allophycocyanin staining and positive enrichment using anti-allophycocyanin MicroBeads (Miltenyi Biotec). Other thymic populations included: DN (lin−CD4−CD8−), DP (CD4+CD8+), and SP (TCRβ+CD4+CD8−) thymocytes. Splenic leukocyte populations included: DCs (CD11c+MHC-II+), T cells (TCRβ+), and B cells (IgM+). BM precursors were defined as Lin−CD117hi cells.
Next-generation sequencing
Messenger RNA from sorted cells was isolated by RNeasy Plus Micro Kit (Qiagen), and then reversely transcribed into complementary DNA using SuperScript II Reverse Transcriptase (Invitrogen). Individual samples for each sequencing run were amplified with primers carrying multiplex identifiers. Sequences are available on request. Amplification was performed using recombinant Taq DNA polymerase (Invitrogen). Amplicons were purified by agarose gel electrophoresis and quantified by Quant-iT dsDNA HS Assay Kit (Invitrogen). Next-generation sequencing was performed on the Genome Sequencer FLX System (454, Roche Applied Sciences) as described before.24
Statistical analysis
All analysis was performed using GraphPad Prism or Excel software. Morisita-Horn index (MHI) and unsupervised hierarchical clustering based on the MHI was calculated based on in-house developed macros.
Results
Canonical early DC progenitors are absent from the adult thymus
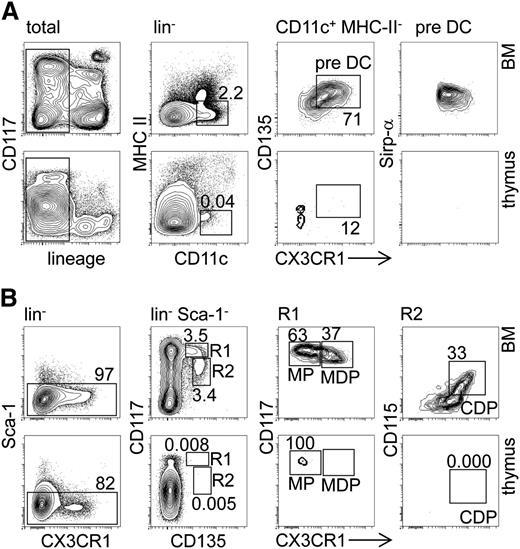
In order to assess whether the thymus contained DC progenitors that were less mature than the previously characterized CD11c+CD207+MHC-IIlo cells,13 such as MPs, MDPs, CDPs, or MHC-II− pre-DCs, we employed CX3CR1GFP mice. MDPs, CDPs, and pre-DCs, which were readily detectable in BM, were completely absent from the thymus (Figure 1A-B). In addition, we found minute numbers of cells phenotypically corresponding to MPs, which might constitute background signals. In conclusion, the adult thymus is virtually devoid of canonical DC progenitors.
Canonical early DC progenitors are absent from the adult thymus. (A) BM (upper panel) and thymic (lower panel) cells from CX3CR1GFP/+ reporter mice were labeled with antibodies against lineage markers (CD19, TCR-β, NK1.1, CD11b, and Gr-1), along with CD11c, MHC-II, and CD135 and analyzed by FACS to detect pre-DCs (CD11c+MHC-II−). Representative FACS density plots indicate the pre-DC population in the BM and thymus. Numbers next to gates indicate the percentage of cells. (B) Similar to (A), the BM and thymic cells from CX3CR1GFP/+ reporter mice were stained with lineage markers (CD19, TCR-β, NK1.1, CD11b, CD11c, and Gr-1), as well as Sca-1, CD135, CD117, and CD115 antibodies to visualize MPs (Lin−Sca-1−CD117hiCD135+CX3CR1−), MDPs (CD117hiCD135+CX3CR1+), and CDPs (CD117+CD135+CX3CR1+CD115+) in the BM (upper panel) and thymus (lower panel). Representative FACS density plots indicate frequencies of MP, MDP, and CDP. Results are representative of 3 independent experiments with at least 3 mice per group.
Canonical early DC progenitors are absent from the adult thymus. (A) BM (upper panel) and thymic (lower panel) cells from CX3CR1GFP/+ reporter mice were labeled with antibodies against lineage markers (CD19, TCR-β, NK1.1, CD11b, and Gr-1), along with CD11c, MHC-II, and CD135 and analyzed by FACS to detect pre-DCs (CD11c+MHC-II−). Representative FACS density plots indicate the pre-DC population in the BM and thymus. Numbers next to gates indicate the percentage of cells. (B) Similar to (A), the BM and thymic cells from CX3CR1GFP/+ reporter mice were stained with lineage markers (CD19, TCR-β, NK1.1, CD11b, CD11c, and Gr-1), as well as Sca-1, CD135, CD117, and CD115 antibodies to visualize MPs (Lin−Sca-1−CD117hiCD135+CX3CR1−), MDPs (CD117hiCD135+CX3CR1+), and CDPs (CD117+CD135+CX3CR1+CD115+) in the BM (upper panel) and thymus (lower panel). Representative FACS density plots indicate frequencies of MP, MDP, and CDP. Results are representative of 3 independent experiments with at least 3 mice per group.
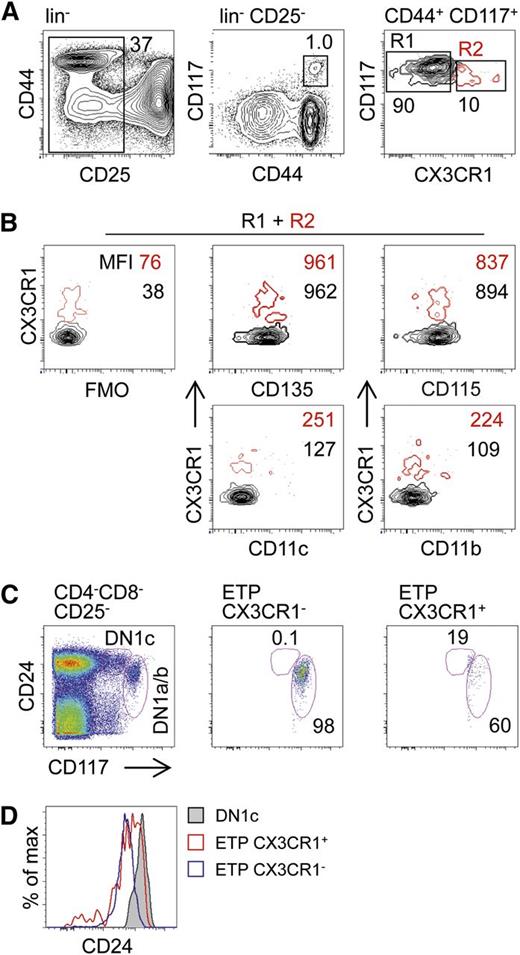
CX3CR1GFP reporter expression is common for MDPs, CDPs, as well as pre-DCs, suggesting that potential noncanonical DC progenitors might also express CX3CR1. Indeed, we identified a subset of CX3CR1+ cells that otherwise phenotypically overlapped with ETPs and constituted approximately 10% of all ETPs (Figure 2A). Further phenotypic characterization for CD135, CD115, CD11c, and CD11b showed no differences between CX3CR1+ and canonical CX3CR1− ETPs (termed ETPs throughout this study) (Figure 2B). Differential expression of CD24 among thymocyte subsets that are considered to be most immature revealed 5 distinct populations designated DN1a-e.25 Within these populations, a subset of DN1c cells, which do not constitute physiological T-lineage progenitors, was identified as a CD207+ DC precursor.13 Analysis of CD24 expression on CX3CR1+ ETPs showed that the majority of these cells were DN1a/b cells, consistent with a function as physiological pro-T cells (Figure 2C). Of note, when compared with canonical CX3CR1− ETPs, CX3CR1+ ETPs expressed slightly lower levels of CD117 and slightly higher levels of CD24, suggesting that these cells might be able to generate DN1c cell progeny (Figure 2C-D). However, the lack of expression of CD11c clearly distinguished CX3CR1+ ETPs from DN1c tDC progenitors.
Identification of ETPs expressing CX3CR1. (A) Thymocytes from CX3CR1GFP/+ reporter mice were depleted of CD4 and CD8 positive cells and subsequently stained with lineage antibodies (CD19, TCR-β, NK1.1, CD11b, CD11c, and Gr-1), as well as CD44, CD25, and CD117. Representative FACS density plots show ETPs (lin−CD44+CD25−CD117hi) negative and positive for CX3CR1. (B) ETPs prepared as in (A) positive (red, R1) or negative (black, R2) for CX3CR1 and were tested for expression of markers associated with myeloid precursors: CD135, CD115, CD11c, and CD11b. FMO controls were used to visualize background fluorescence. (C) Thymocytes were prepared and stained as in (A) and antibodies against CD24 to discriminate DN1 subsets according to the DN1a-e scheme. (D) Levels of CD24 on DN1 cells. Numbers inside contour plots indicate median fluorescent intensities of the respective surface antigens. Data are representative of 6 (A) and 2 (B-D) independent experiments. FMO, fluorescence-minus-one.
Identification of ETPs expressing CX3CR1. (A) Thymocytes from CX3CR1GFP/+ reporter mice were depleted of CD4 and CD8 positive cells and subsequently stained with lineage antibodies (CD19, TCR-β, NK1.1, CD11b, CD11c, and Gr-1), as well as CD44, CD25, and CD117. Representative FACS density plots show ETPs (lin−CD44+CD25−CD117hi) negative and positive for CX3CR1. (B) ETPs prepared as in (A) positive (red, R1) or negative (black, R2) for CX3CR1 and were tested for expression of markers associated with myeloid precursors: CD135, CD115, CD11c, and CD11b. FMO controls were used to visualize background fluorescence. (C) Thymocytes were prepared and stained as in (A) and antibodies against CD24 to discriminate DN1 subsets according to the DN1a-e scheme. (D) Levels of CD24 on DN1 cells. Numbers inside contour plots indicate median fluorescent intensities of the respective surface antigens. Data are representative of 6 (A) and 2 (B-D) independent experiments. FMO, fluorescence-minus-one.
CX3CR1+ and CX3CR1− ETPs have comparable DC potential in vitro
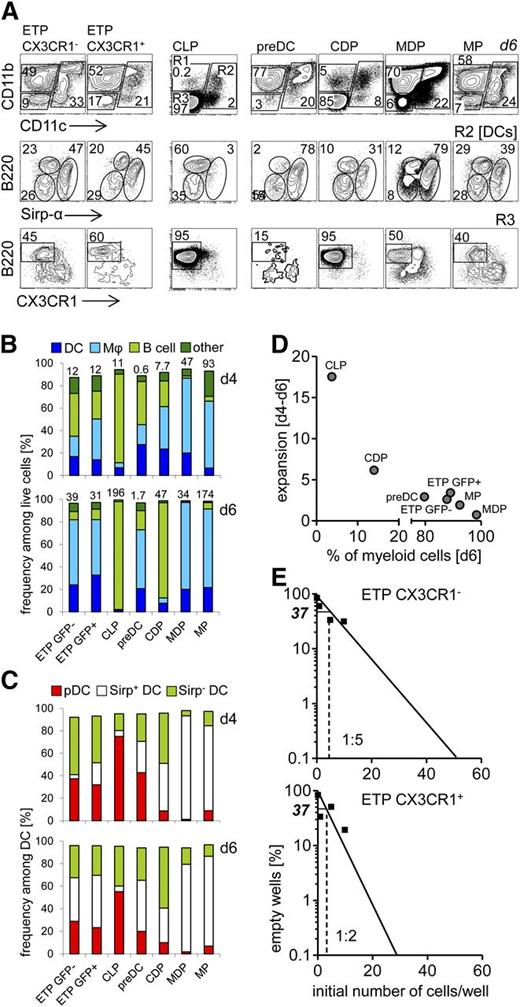
Next, we employed the OP9 coculture system to compare the propensity of CX3CR1+ ETPs to differentiate into non-T cells with that of their CX3CR1− counterparts, as well as other canonical lymphoid and DC progenitor populations.26 After 4 days of culture CX3CR1+ ETPs, ETPs, pre-DCs, and CDPs displayed a very similar distribution of DC, macrophage, and B-lineage progeny (Figure 3A-B) with ETPs generating slightly fewer Sirpα+ DCs and CDPs generating only a few pDCs (Figure 3C). After 6 days of culture, ratios of progeny were shifted toward myeloid populations in cultures of CX3CR1+ ETPs, ETPs, and pre-DCs. In contrast, CDP-derived cultures consisted predominantly of B cells, whereas MPs and MDPs generated mostly DCs and macrophages, and the majority of DCs were Sirpα+ (Figure 3B-C). Common lymphoid progenitors (CLPs) predominantly gave rise to B cells, and limited but detectable numbers of macrophages and DCs, the latter consisting mostly of pDCs and very limited numbers of Sirpα+ DCs (Figure 3C).
CX3CR1+ and CX3CR1− ETPs have comparable DC potential in vitro. (A) BM-derived progenitors (CLPs, pre-DCs, CDPs, MDPs, and MPs) and thymic progenitors (CX3CR1+ and CX3CR1− ETPs) were sorted from CX3CR1GFP/+ reporter mice. Progenitors were sorted according to gates shown in Figures 1 and 2. CLPs were defined as lin−Sca-1+CD117intCD127+ CD135+. Purified precursors were cultured on OP9 stromal cells in the presence of Flt-3L, SCF, IL-7, and GM-CSF, and analyzed at days 4 and 6 to assess their myeloid and B-lineage potential. Cells derived from these precursors were defined as DC, based on expression of CD11c, along with Sirpα and B220. Bona fide macrophages were identified as CD11b+CD11c−, while B220 cells were defined as CD11c−CD11b−B220+CX3CR1−. (B) Quantification of data shown in (A) at day 4 (upper panel) and day 6 (bottom panel) of culture. Numbers above the columns indicate fold expansion of precursors compared with day 0. (C) Quantification of DC populations shown in (A, middle panel) at day 4 (upper panel) and day 6 (bottom panel) of culture. Development of 3 major DC populations was assessed: CD11c+Sirpα+, CD11c+Sirpα−, and CD11c+B220+ (pDC). (D) Expansion of precursors between days 4 to 6 inversely correlated to the frequency of myeloid cells generated by day 6. (E) The potential of CX3CR1+ and CX3CR1− ETPs to develop toward the myeloid or B lineages was assessed by limiting dilution assay on OP9 stromal cells in the presence of Flt-3L, SCF, IL-7, and GM-CSF. (A-E) Data are representative of 2 independent experiments.
CX3CR1+ and CX3CR1− ETPs have comparable DC potential in vitro. (A) BM-derived progenitors (CLPs, pre-DCs, CDPs, MDPs, and MPs) and thymic progenitors (CX3CR1+ and CX3CR1− ETPs) were sorted from CX3CR1GFP/+ reporter mice. Progenitors were sorted according to gates shown in Figures 1 and 2. CLPs were defined as lin−Sca-1+CD117intCD127+ CD135+. Purified precursors were cultured on OP9 stromal cells in the presence of Flt-3L, SCF, IL-7, and GM-CSF, and analyzed at days 4 and 6 to assess their myeloid and B-lineage potential. Cells derived from these precursors were defined as DC, based on expression of CD11c, along with Sirpα and B220. Bona fide macrophages were identified as CD11b+CD11c−, while B220 cells were defined as CD11c−CD11b−B220+CX3CR1−. (B) Quantification of data shown in (A) at day 4 (upper panel) and day 6 (bottom panel) of culture. Numbers above the columns indicate fold expansion of precursors compared with day 0. (C) Quantification of DC populations shown in (A, middle panel) at day 4 (upper panel) and day 6 (bottom panel) of culture. Development of 3 major DC populations was assessed: CD11c+Sirpα+, CD11c+Sirpα−, and CD11c+B220+ (pDC). (D) Expansion of precursors between days 4 to 6 inversely correlated to the frequency of myeloid cells generated by day 6. (E) The potential of CX3CR1+ and CX3CR1− ETPs to develop toward the myeloid or B lineages was assessed by limiting dilution assay on OP9 stromal cells in the presence of Flt-3L, SCF, IL-7, and GM-CSF. (A-E) Data are representative of 2 independent experiments.
Of note, cultures originating from the various progenitors expanded to different extents during differentiation (Figure 3B). MPs and MDPs displayed the strongest expansion within 4 days of culture, followed by CDPs, CLPs, and ETP subsets, all of which expanded approximately 10-fold. In contrast, pre-DCs failed to expand during this interval. After 6 days of culture, MP- and CLP-derived cultures had expanded close to 200-fold, whereas CDP and ETP-derived cultures had expanded 30- to 50-fold. Notably, no additional expansion was observed in MDP- and pre–DC-derived cultures after 6 days when compared with 4 days of culture. Whereas early expansion is likely to indicate more the potential of the input progenitors to proliferate and/or survive under these culture conditions, further expansion between days 4 and 6 of culture inversely correlated with the frequency of myeloid cells in these cultures, consistent with their state of terminal differentiation (Figure 3D). Comparison of CX3CR1+ ETPs and ETPs in limiting dilution analysis revealed a slightly higher precursor frequency within the ETPs in OP9 cocultures (Figure 3E). In conclusion, all progenitors, including CLPs, displayed myeloid potential in vitro. In turn, somewhat unexpectedly, B-lineage potential, albeit to different degrees, was found in all populations as well. In addition, limiting dilution analysis showed that CX3CR1+ ETPs contained a higher frequency of myeloid-lineage progenitors when compared with their canonical counterparts.
T-lineage differentiation can be elicited from various myeloid lineage-committed or DC-committed progenitors
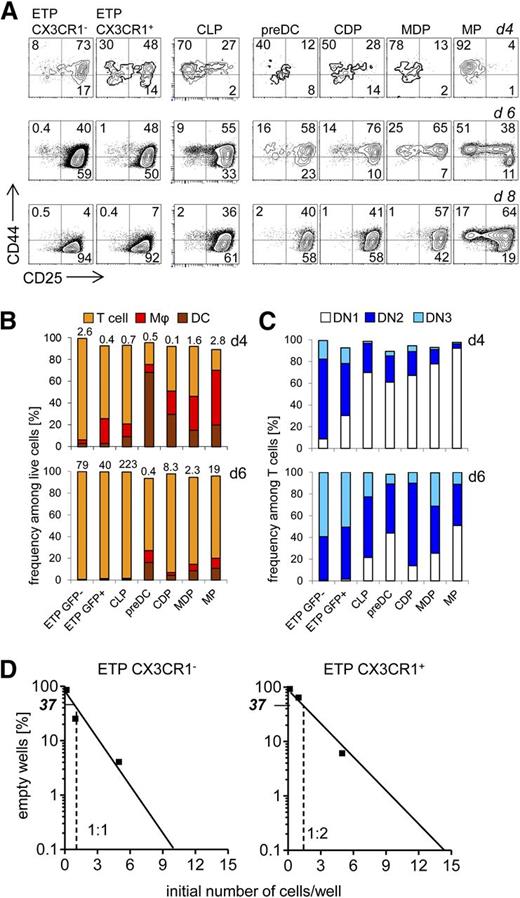
Given the lymphoid lineage potential of DC progenitor populations detected in vitro, we next assessed whether these populations carried the potential to differentiate into T-lineage cells as well. To this end, we employed the OP9-DL1 coculture system.20 CLPs and both ETP subsets gave rise predominantly to T-lineage cells after 6 days of culture (Figure 4A-B). Consistent with previous work,27 ETPs displayed a more rapid developmental progression when compared with CLPs (Figure 4A,C). Both subpopulations of ETPs displayed a similar T-lineage progenitor frequency in limiting dilution analysis with a higher frequency in ETPs (Figure 4D). As expected, MPs yielded mostly DC-like and macrophage-like cells at this time point, but we could also detect some CD25+ DN2 cells in each culture (supplemental Figure 2 and Figure 4A-B). Of note, after 6 days of culture, T-lineage cells constituted the majority irrespective of the starting population and most of these cells displayed a CD25+ DN2 or DN3 phenotype (Figure 4A,C). However, cultures from canonical lymphoid progenitors, including CX3CR1+ ETPs, expanded 40- to 223-fold. In contrast, cultures from myeloid-lineage progenitors (with the exception of MP-derived cultures) expanded considerably less (Figure 4B). Pre–DC-derived cultures even underwent a twofold contraction during the culture period of 6 days. Taken together, we conclude that all DC progenitor populations tested, ranging from MPs to pre-DCs, have robust T-lineage potential in vitro. These data suggest that inductive signals from the environment may critically contribute to DC vs T-lineage fate decisions in the thymus.
T-lineage differentiation can be elicited from various myeloid–lineage-committed or DC-committed progenitors. (A) BM-derived and thymic progenitors were sorted from CX3CR1GFP/+ reporter mice. Purified precursors were cultured on OP9-DL1 stromal cells in the presence of Flt-3L, SCF, and IL-7, and analyzed at days 4, 6, and 8 to assess their myeloid- and T-lineage potential. DCs were considered as CD11c+, while macrophages were CD11b+CD11c−. T-cell–committed precursors were negative for myeloid markers (CD11c, CD11b, and CX3CR1) and further subdivided into 3 DN populations based on the surface expression of CD25 and CD44. DN1 were CD44+CD25−, DN2 CD44+CD25+, and DN3 CD44−CD25+. (B) Quantification of data shown in (A) at day 4 (upper panel) and day 6 (bottom panel) of culture. Numbers above the columns indicate fold expansion of precursors. (C) Quantification of T-lineage–committed DN populations shown in (A) at day 4 (upper panel) and day 6 (bottom panel) of culture. (D) The potential of CX3CR1+ and CX3CR1− ETPs to develop toward the T lineage was assessed by limiting dilution assay on OP9-DL1 stromal cells in the presence of Flt-3L, SCF, and IL-7. (A-D) Data are representative of 2 independent experiments.
T-lineage differentiation can be elicited from various myeloid–lineage-committed or DC-committed progenitors. (A) BM-derived and thymic progenitors were sorted from CX3CR1GFP/+ reporter mice. Purified precursors were cultured on OP9-DL1 stromal cells in the presence of Flt-3L, SCF, and IL-7, and analyzed at days 4, 6, and 8 to assess their myeloid- and T-lineage potential. DCs were considered as CD11c+, while macrophages were CD11b+CD11c−. T-cell–committed precursors were negative for myeloid markers (CD11c, CD11b, and CX3CR1) and further subdivided into 3 DN populations based on the surface expression of CD25 and CD44. DN1 were CD44+CD25−, DN2 CD44+CD25+, and DN3 CD44−CD25+. (B) Quantification of data shown in (A) at day 4 (upper panel) and day 6 (bottom panel) of culture. Numbers above the columns indicate fold expansion of precursors. (C) Quantification of T-lineage–committed DN populations shown in (A) at day 4 (upper panel) and day 6 (bottom panel) of culture. (D) The potential of CX3CR1+ and CX3CR1− ETPs to develop toward the T lineage was assessed by limiting dilution assay on OP9-DL1 stromal cells in the presence of Flt-3L, SCF, and IL-7. (A-D) Data are representative of 2 independent experiments.
Limited lineage relationship of tDCs and thymocytes revealed by retroviral barcoding
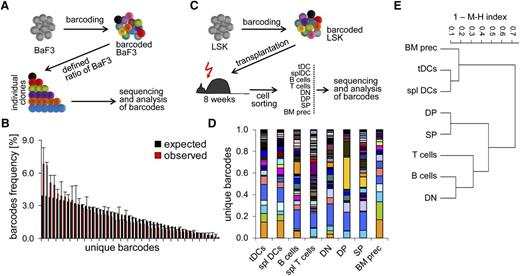
In order to test in vivo whether tDCs and thymocytes are developmentally related, we employed retroviral barcoding combined with next-generation sequencing as an independent approach to determine potential lineage relationships. We hypothesized that limited colonization of the thymus by BM-derived progenitors would allow us to determine whether thymocytes and tDCs shared a common intrathymic progenitor. Barcodes consisted of semi-random oligonucleotide duplexes, in which 6 pairs of random nucleotides were separated by fixed triplets, with a theoretical diversity of more than 106 (supplemental Figure 1A).28
In order to gain an estimate on the sensitivity of our barcoding approach, we transduced the lymphoid cell line BaF3 and generated 50 clones carrying defined individual barcodes. These clones were then mixed at a defined ratio of 50:49:48:…:3:2:1 into one single culture (Figure 5A). After 3 days of culture, 50 000 cells were randomly harvested from this culture and subjected to next-generation sequencing. Comparison of the expected frequencies of barcodes ranging from 3.9% to 0.08% for the most and the least abundant ones, respectively, with the barcodes recovered from the sequencing experiment showed that our system is sufficiently sensitive to faithfully detect small differences in clone sizes and to recover clones of very low abundance (Figure 5B).
Limited lineage relationship of tDCs and thymocytes revealed by retroviral barcoding. (A) Experimental setup. Fifty individually barcoded BaF3 cells were clonally expanded in vitro. Bar coded BaF3 clones were then mixed at defined ratios of 50:49:48 … 3:2:1 and 50 000 cells of these mixed cultures were subjected to bar code analysis by 454 sequencing. (B) Distribution of individual barcode sequences in BaF3 test sample. Expected frequencies of individual bar codes are shown in black. Observed frequencies of individual barcodes after sequencing are displayed in red. (C) Experimental setup to assess lineage relationship between tDCs, splenic DCs, and thymocytes. LSK cells were transduced with retroviral vectors encoding the barcode library at a multiplicity of infection, on average generating a single integration event per cell. Tagged LSK were FACS-sorted based on GFP expression and injected into lethally irradiated recipients to generate BM chimeras. About 8 to 10 weeks after transplantation, indicated populations of cells were sorted (50 000 cells) and sequenced as described in (A). (D) The distribution of unique barcodes was assessed for tDCs, splenic DCs, splenic B and T cells, DN, DP, and SP thymocytes, as well as BM lin−CD117+ precursors. Each color represents a unique barcode. (E) Unsupervised hierarchical clustering based on the MHI to reveal lineage relationships between sorted populations. (B,D-E) Data are pooled of 2 independent experiments with a total number of 6 mice analyzed.
Limited lineage relationship of tDCs and thymocytes revealed by retroviral barcoding. (A) Experimental setup. Fifty individually barcoded BaF3 cells were clonally expanded in vitro. Bar coded BaF3 clones were then mixed at defined ratios of 50:49:48 … 3:2:1 and 50 000 cells of these mixed cultures were subjected to bar code analysis by 454 sequencing. (B) Distribution of individual barcode sequences in BaF3 test sample. Expected frequencies of individual bar codes are shown in black. Observed frequencies of individual barcodes after sequencing are displayed in red. (C) Experimental setup to assess lineage relationship between tDCs, splenic DCs, and thymocytes. LSK cells were transduced with retroviral vectors encoding the barcode library at a multiplicity of infection, on average generating a single integration event per cell. Tagged LSK were FACS-sorted based on GFP expression and injected into lethally irradiated recipients to generate BM chimeras. About 8 to 10 weeks after transplantation, indicated populations of cells were sorted (50 000 cells) and sequenced as described in (A). (D) The distribution of unique barcodes was assessed for tDCs, splenic DCs, splenic B and T cells, DN, DP, and SP thymocytes, as well as BM lin−CD117+ precursors. Each color represents a unique barcode. (E) Unsupervised hierarchical clustering based on the MHI to reveal lineage relationships between sorted populations. (B,D-E) Data are pooled of 2 independent experiments with a total number of 6 mice analyzed.
To determine lineage relationships in vivo, LSK cells were transduced so that the vast majority of cells contained one individual barcode.29 Barcoded cells were sorted based on eGFP expression and used to generate BM chimeras. After 8 weeks, complementary DNA from fluorescence-activated cell sorter (FACS)-sorted tDCs, splenic DCs, B and T cells, DN, DP, and SP thymocytes, as well as BM-derived Lin−CD117+ precursors was subjected to next-generation sequencing (Figure 5C). Analysis of the frequency of individual recovered barcodes revealed a high similarity between tDCs, splenic DCs, and BM-derived progenitors. In addition, DP and SP thymocytes had a comparable distribution of bar codes, which was clearly distinct from that of DCs (Figure 5D). The MHI is a measure of similarity, which scores identical populations as 1 and completely distinct populations as 0.30 In order to quantify clonal relationships between sorted populations, we compared each possible pair of populations and subjected the resulting MHIs to unsupervised hierarchical clustering (Figure 5E). These calculations substantiated the initial impression of a close lineage relationship between DCs, irrespective of their origin, as well as BM-derived progenitors (Figure 5E). In contrast, all lymphoid populations were distinct from those 3 subsets. The closest linkage was observed between DP and SP thymocytes, consistent with a close temporal developmental relationship. DN thymocytes were less related to other thymocytes, possibly reflecting a heterogeneous mixture of populations undergoing dynamic changes during its lifespan. Interestingly, DN thymocytes were most closely related to splenic B cells, which might reflect their simultaneous generation from the same small population of common progenitors. In conclusion, these data indicate that a large majority of tDCs do not arise from the same intrathymic progenitor as T cells. Rather, tDCs and splenic DCs are likely to share a common origin.
Generation of a niche for DCs permits intrathymic DC development
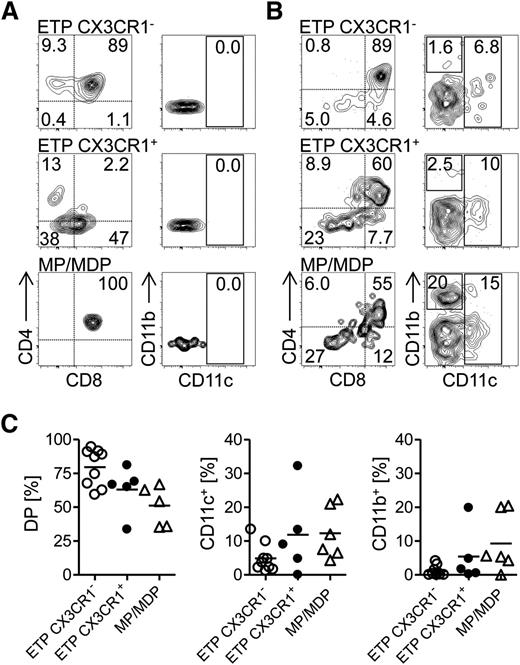
Our data suggested that limited lineage relationships between tDCs and thymocytes in vivo despite multipotentiality of progenitors might be due to strong inductive signals, such as via Notch ligands in OP9-DL1 cultures. Therefore, we next addressed the question whether the thymic microenvironment transmits such inductive signals and whether it can be manipulated toward being permissive to non–T-lineage differentiation. To this end, we transferred congenically marked (CD45.1) ETPs, CX3CR1+ ETPs, or mixed MP/MDP cells directly into the thymus of nonmanipulated mice and assessed their capacity to generate T-lineage or DC-lineage progeny after 21 days. ETPs readily gave rise to DP thymocytes, whereas CX3CR1+ ETPs generated much fewer DP thymocytes (Figure 6A). None of the two populations generated DC-lineage progeny under these conditions (Figure 6A). Interestingly, transferred MP/MDP also gave rise to T-lineage cells, but not DC-lineage cells under these conditions, suggesting that the nonmanipulated thymus transmits strong inductive signals to drive progenitor cells into the T lineage.
Generation of a niche for DCs permits intrathymic DC development. (A) Sorted thymic CX3CR1+ ETPs, CX3CR1− ETPs, and BM MP/MDP precursors were injected intrathymically into congenic (CD45.1+CD45.2+), nonmanipulated mice. The development of myeloid cells and T cells was assessed 21 days after transfer. CD45.2+ T-cell–committed precursors were either DP or SP for CD4 and CD8. Myeloid cells were CD4/CD8 DN and expressed CD11c (DC), or were CD11c−CD11b+ (macrophages). (B) Precursors sorted as in (A) were injected intrathymically into CD11c-DOG (CD45.1+CD45.2+) mice, in which DCs were depleted 24 hours prior to progenitor injection. Development of T cells and myeloid cells was assessed 21 days after transfer. (C) Quantification of data shown in (B). (A-B) Representative plots of 2 independent experiments and (C) pooled data of 2 independent experiments, with each point representing an individual mouse.
Generation of a niche for DCs permits intrathymic DC development. (A) Sorted thymic CX3CR1+ ETPs, CX3CR1− ETPs, and BM MP/MDP precursors were injected intrathymically into congenic (CD45.1+CD45.2+), nonmanipulated mice. The development of myeloid cells and T cells was assessed 21 days after transfer. CD45.2+ T-cell–committed precursors were either DP or SP for CD4 and CD8. Myeloid cells were CD4/CD8 DN and expressed CD11c (DC), or were CD11c−CD11b+ (macrophages). (B) Precursors sorted as in (A) were injected intrathymically into CD11c-DOG (CD45.1+CD45.2+) mice, in which DCs were depleted 24 hours prior to progenitor injection. Development of T cells and myeloid cells was assessed 21 days after transfer. (C) Quantification of data shown in (B). (A-B) Representative plots of 2 independent experiments and (C) pooled data of 2 independent experiments, with each point representing an individual mouse.
Next, we tested whether it was possible to induce intrathymic DC differentiation from different progenitors by altering the thymic microenvironment. To this end, we employed CD11c-DOG mice where DCs can be depleted by injection of DT.18 ETPs, CX3CR1+ ETPs, or MP/MDP from congenic (CD45.1) donors were transferred intrathymically into DC-depleted CD11c-DOG mice and their progeny was analyzed after 21 days. Similar to transfers into nonmanipulated recipients, all donor populations gave rise to T-lineage progeny (Figure 6B). Notably, the frequencies of donor-derived DP thymocytes in DC-depleted animals were higher after transfer of CX3CR1+ ETPs and MP/MDPs than after transfer into nonmanipulated recipients, suggesting that these cells are initially dependent on niches occupied by DCs (Figure 6C). In addition, in DC-depleted mice, all donor populations gave rise to CD11c+, as well as CD11b+ myeloid progeny with ETPs, showing a reduced capacity to generate myeloid cells when compared with CX3CR1+ ETPs and MP/MDP cells (Figure 6B-C). Thus, the depletion of DCs generates a permissive state for myelopoiesis and DC differentiation in the thymus.
Discussion
The origin of thymus-resident cDCs, as well as that of other myeloid cells in the thymus has been controversially discussed. Experimental evidence exists that supports both common and separate origins as thymocytes. Although some of these conflicting data may be explained by high sensitivity of stromal cocultures to elicit non–T-lineage fate from T-cell progenitors (possibly due to limited availability of Notch ligands),14,31 others are more difficult to explain. Synchronized intrathymic development of tDCs and thymocytes might be explained by a general gate opening process generating a permissive state for thymus colonization by progenitor cells irrespective of their developmental potential.32 A pre-thymic transient convergence of developmental pathways appears to be required to explain immunoglobulin rearrangements in thymic myeloid cells.14 However, why only thymic myeloid cells should carry these rearrangements in such a scenario is difficult to fathom.
Identification of a common DC–T-cell progenitor would constitute a prerequisite if at least some tDCs and thymocytes shared a common intrathymic origin. Because canonical DC precursors were absent from the thymus, we focused on the characterization of a subset of ETPs expressing CX3CR1, which can be described as a hallmark of DC progenitors. Ten percent of all ETPs were CX3CR1GFP positive, a frequency consistent with that showing no history of Il7r expression in a previous fate mapping study.11 This subset displayed a marginally higher precursor frequency in non–T-lineage promoting limiting dilution assays, as well as a slightly higher capacity to generate tDCs in DC-depleted animals when compared with CX3CR1− ETPs. Given the apparent absence of ETP-derived development of tDCs in vivo, CX3CR1 expression might reflect a state of aborted lineage diversion at steady state.
Viral barcoding has emerged as a new technology to retrospectively track the behavior of cells at the clonal level. This approach has been proven useful to track differential clonal expansion of T cells during an immune response, as well as to determine hematopoietic lineage relationships.28,33-35 Here, we have employed this method to assess lineage relationships between tDCs and thymocytes in the thymus. This experiment was based on the assumption that thymus colonization represents a bottle neck in T-cell development,36 and that, therefore, thymocytes carry a limited set of barcodes, which is distinct from all other hematopoietic subsets. Our data showed that tDCs and splenic cDCs were closely related, whereas all T-cell subsets analyzed clustered together and displayed little similarity in terms of barcodes when compared with either set of DCs. These data support the notion that tDCs and thymocytes do not share an intrathymic progenitor, although several caveats remain to be considered. It constitutes one caveat that because of low cell numbers amenable to analysis, heterogeneous populations had to be subjected to sequencing. Thus, the analysis of total conventional tDCs rather than CD8α+ DCs might result in an underestimation of a putative thymocyte-DC relationship, because only these DCs are thought to be generated in situ.37 Furthermore, DN thymocytes combined might contain cells originating from sequential colonization events, as thymocytes remain DN for up to 18 days.38 This notion is consistent with our finding that total DN cells were overall more closely related to B cells than to the more mature DP and SP thymocytes, which in part have already undergone selection events. Thus, the similarity in barcode distribution between DN thymocytes and B cells possibly reflects their recent common origin from a lymphoid progenitor. Ultimately, we cannot completely exclude that the observed difference between barcodes recovered from tDCs and DN thymocytes is also caused by selection of a minor subset of DN cells to develop into tDCs. However, such a scenario would still not explain the high degree of similarity observed between splenic DCs and tDCs. Taking into account these caveats, the high degree of dissimilarity of all DCs to thymocytes and the closer relationships of DN thymocytes to more mature thymocyte lineages, as well as peripheral T and even B cells, nevertheless provide no indication for a common tDC-thymocyte progenitor.
Depletion of endogenous DCs using DT generated a permissive state for intrathymic DC development originating from various progenitors, including MP/MDPs and ETPs. These data are consistent with a previous report that showed that ablation of Notch1 in T-cell progenitors allowed their diversion into the DC lineage.16 This study suggested that the thymic microenvironment provided inductive signals via Dll4-Notch1 interactions to prevent lineage diversion of T-lineage progenitors. Our study indicates that even wild-type progenitors are able to escape inductive signaling, once space, which is otherwise occupied by tDCs, is provided. The nature of a putative niche devoid of Dll4 yet remains elusive. However, the absence of DCs might result in high local concentrations of Flt3-L promoting intrathymic DC reconstitution. This hypothesis is consistent with the role of Flt3-L as a rheostat for DC homeostasis in the periphery.18 Given the absence of early myeloid and uncommitted DC progenitors in the thymus, empty tDC niches could potentially be filled by ETPs provided they escape Notch signaling. Genetic ablation of Notch1 or depletion of DCs constitute massive alterations of the thymic microenvironment, which might, similar to highly sensitive in vitro assays, reveal developmental potential that might not be elicited in a physiological situation at steady state.14,31 Consistently, previous work based on fate mapping, as well as our barcoding approach, suggest that T-lineage progenitors generating tDCs might be a rare event at best.11 Rather, empty tDC niches are likely to be filled by dedicated committed intrathymic progenitors, such as DN1c cells, although their prethymic origins remain to be characterized.13
In conclusion, our study supports a model in which the majority of thymus-resident DCs are generated from already committed progenitors, whereas noncommitted progenitors are forced into the T lineage due to strong inductive signals from the thymic microenvironment. These findings are consistent with recent fate mapping experiments.11,12,16 Liberation of niches occupied by tDCs generates a permissive state for T-lineage progenitors to adopt an alternative fate, although the extent to which this alternative pathway of DC development operates in different physiological scenarios remains to be investigated. Of note, the presence of immunoglobulin gene rearrangements exclusively in thymic but not in other cDC subsets suggests, at least in some instances, that tDCs might be generated through this alternative pathway. Taken together, our study underscores the plasticity of the thymus to support the development of a balanced pool of cell types that is essential for its function.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Reinhold Förster for critical reading of the manuscript. The authors acknowledge the assistance of the Cell Sorting Core Facility of the Hannover Medical School, supported in part by Braukmann-Wittenberg-Herz-Stiftung and the German Research Foundation. The CD11c.DOG mice were kindly provided by Natalio Garbi (University of Bonn, Germany).
This study was supported by grants from the German Research Foundation (Emmy-Noether Program; KR2320/2-1, SFB738-A7, KR2320/3-1, and EXC62 “Rebirth”) (A.K.) and SFB738-C9 (A.S.).
Authorship
Contribution: M.Ł., N.Z., A.S., and A.K. designed research; M.Ł., N.Z., L.F., J.D., K.W., and A.S. performed research; M.Ł., N.Z., L.F., J.P., I.P., and A.K. analyzed data; and M.Ł. and A.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andreas Krueger, Institute of Immunology, OE5240, Hannover Medical School, Carl-Neuberg-Strasse 1, 30625 Hannover, Germany; e-mail: krueger.andreas@mh-hannover.de.