Abstract
Intrachromosomal amplification of chromosome 21 (iAMP21) defines a distinct cytogenetic subgroup of childhood B-cell precursor acute lymphoblastic leukemia. Breakage-fusion-bridge cycles followed by chromothripsis and other complex structural rearrangements of chromosome 21 underlie the mechanism giving rise to iAMP21. Patients with iAMP21 are older (median age 9 years), with a low white cell count. They have a high relapse rate when treated as standard risk. Recent studies have shown improved outcome on intensive therapy. Molecular targets for therapy are being sought.
In childhood acute lymphoblastic leukemia (ALL), chromosomal abnormalities define specific cytogenetic subgroups with distinctive clinical features. Of greatest significance is their association with outcome; well-known examples include the good prognosis associated with the translocation t(12;21)(p13;q22)/ETV6-RUNX1 fusion and the poor outcome linked to the presence of the Philadelphia chromosome.1 Such abnormalities are now routinely used in risk stratification for treatment, which has made a substantial contribution to the remarkably improved outcome of 90% event-free survival (EFS) for childhood B-cell precursor ALL (BCP-ALL). Intrachromosomal amplification of chromosome 21 (iAMP21) is a more recently identified chromosomal abnormality in childhood BCP-ALL, which has strong prognostic associations. Study of the processes leading to the formation of the iAMP21 chromosome has elucidated novel mechanisms important in leukemogenesis, at least in this subtype, but likely with wider implications.
Cytogenetic definition of iAMP21
iAMP21 was identified as a distinct cytogenetic subgroup of BCP-ALL in 2003,2,3 following reports of a number of sporadic cases. It was detected during routine screening for the presence of the ETV6-RUNX1 fusion by fluorescence in situ hybridization (FISH). In the absence of the fusion, approximately 2% of childhood BCP-ALL patients showed additional copies of signals specific for the RUNX1 gene. The signals were seen in tandem duplication along the length of a grossly abnormal chromosome 21 (the iAMP21 chromosome) in metaphase and clustered together in interphase2 (Figure 1). From cytogenetic analysis, the morphology of the abnormal chromosome 21 varied markedly between patients, which is likely why it had not been previously described. Rare cases have been seen in which iAMP21 occurs in association with other established chromosomal changes, including high hyperdiploidy, BCR-ABL1, or ETV6-RUNX14-6 ; otherwise, it has been confirmed to be a primary cytogenetic change, which remains constant in structure between diagnosis and relapse.7 Similar abnormalities of chromosome 21 have been rarely reported in acute myeloid leukemia and myelodysplastic syndromes, usually in association with complex karyotypes.8,9 However, the chromosomal regions involved in such cases appear to be different.
Examples of FISH and next-generation sequencing iAMP21 data. (A) Cells from a patient with iAMP21. The red signals indicate copies of RUNX1, and the green signals indicate copies of ETV6 using FISH with a dual-color probe for the detection of the ETV6-RUNX1 fusion. No fusions are present in these cells. In the metaphase, the 2 pairs of green signals indicate the normal location of ETV6 on the short arm of the 2 normal chromosome 12 homologs. The individual pair of red signals on the right-hand side of the cell shows the location of RUNX1 on the normal chromosome 21. On the left-hand side of the cell, the cluster of red signals indicates amplification of RUNX1 on the abnormal chromosome 21 (iAMP21 chromosome). In the interphase cell (inset), the 2 green signals indicate the presence of 2 copies of ETV6. The cluster of red signals indicates the amplification of RUNX1. The fact that these signals are clustered implies that they are located in close proximity on a single chromosome. These characteristics define iAMP21. (B) A typical sporadic-iAMP21 copy-number profile of the chromosome 21 derived from paired-end sequencing data.21 Fluctuations above and below a copy number of 2 indicate gain and deletion, respectively. The profile covers the whole of chromosome 21, from centromere (left had side) to telomere (right hand side), and the chromosome positions are indicated in Mb. The deletion of the telomeric region indicates the breakpoint from which the BFB cycle was initiated in this patient. The stepwise changes in copy number along chromosome 21 represent the typical profile generated from BFB cycles, with the most highly amplified region juxtaposed to the deleted telomeric region. There are a small number of rearrangements resulting from chromothripsis in this patient. The originally defined common region of amplification of 5.1 Mb is indicated by the red bar, and the consistently most highly amplified and overexpressed regions generated from a consensus copy-number profile21 are indicated by the green bars. These regions include genes important in hematologic malignancies, and some examples with their relative locations are shown in the red box.
Examples of FISH and next-generation sequencing iAMP21 data. (A) Cells from a patient with iAMP21. The red signals indicate copies of RUNX1, and the green signals indicate copies of ETV6 using FISH with a dual-color probe for the detection of the ETV6-RUNX1 fusion. No fusions are present in these cells. In the metaphase, the 2 pairs of green signals indicate the normal location of ETV6 on the short arm of the 2 normal chromosome 12 homologs. The individual pair of red signals on the right-hand side of the cell shows the location of RUNX1 on the normal chromosome 21. On the left-hand side of the cell, the cluster of red signals indicates amplification of RUNX1 on the abnormal chromosome 21 (iAMP21 chromosome). In the interphase cell (inset), the 2 green signals indicate the presence of 2 copies of ETV6. The cluster of red signals indicates the amplification of RUNX1. The fact that these signals are clustered implies that they are located in close proximity on a single chromosome. These characteristics define iAMP21. (B) A typical sporadic-iAMP21 copy-number profile of the chromosome 21 derived from paired-end sequencing data.21 Fluctuations above and below a copy number of 2 indicate gain and deletion, respectively. The profile covers the whole of chromosome 21, from centromere (left had side) to telomere (right hand side), and the chromosome positions are indicated in Mb. The deletion of the telomeric region indicates the breakpoint from which the BFB cycle was initiated in this patient. The stepwise changes in copy number along chromosome 21 represent the typical profile generated from BFB cycles, with the most highly amplified region juxtaposed to the deleted telomeric region. There are a small number of rearrangements resulting from chromothripsis in this patient. The originally defined common region of amplification of 5.1 Mb is indicated by the red bar, and the consistently most highly amplified and overexpressed regions generated from a consensus copy-number profile21 are indicated by the green bars. These regions include genes important in hematologic malignancies, and some examples with their relative locations are shown in the red box.
The complexity and variability of the iAMP21 chromosome in BCP-ALL, comprising multiple regions of gain, amplification, inversion, and deletion, were further highlighted by FISH and genomic analysis.7,10,11 However, in spite of the differences in their genomic profiles, consistent characteristics exist among iAMP21 patients. These features include a common region of highest-level amplification spanning 5.1 Mb of chromosome 21 from 32.8 to 37.9 Mb, within which the RUNX1 gene is located, combined with the lowest-level copy number distal of this region to the telomere7 (Figure 1). These observations have confirmed that FISH, using probes directed to RUNX1, to determine the number of copies of the most highly amplified region, provides a reliable detection method.12 Thus, the finding of 3 or more extra copies of RUNX1 on a single abnormal chromosome 21 (a total of 5 or more RUNX1 signals per cell) is currently used as the international definition of iAMP2113 (Figure 1). Interpretation of cases with interphase cells only should be made with caution, as extra RUNX1 signals will also define additional copies of chromosome 21, which are characteristic of high-hyperdiploid ALL. As a result of such concerns, more recently, the distinctive genomic profile of chromosome 21 is being used to confirm the accuracy of iAMP21 diagnosis (Figure 1).
Although RUNX1 has been highlighted by its location within the highly amplified region of the chromosome and its use as a marker to define iAMP21, there is no evidence as yet that it is the target gene of this abnormality. RUNX1 was found not to be differentially expressed among gene expression profiles of childhood ALL patients, when samples from iAMP21 patients were specifically compared with samples from BCP-ALL patients with additional copies or other rearrangements of chromosome 21 (high hyperdiploidy and ETV6-RUNX1 with additional copies of the derivative chromosome 21, respectively).11 In addition, no mutations of RUNX1 coding exons, and specifically those within the Runt domain (exons 3, 4, and 5), have been found.7,14,15 With good reason, RUNX1 is a strong candidate for a driver of the leukemic process in iAMP21 patients, but in the absence of evidence, other genes within the amplified region and or rearranged within the abnormal chromosome 21 have equal opportunity.
Patients with iAMP21 display a unique spectrum of secondary genetic abnormalities, likely contributing to disease progression, which may also be used for improved diagnosis. These include gain of chromosomes X, 10, or 14; or monosomy 7/deletion of 7q; deletions of 11q, including the ATM and MLL genes P2RY8-CRLF2; and deletions of ETV6 and RB1.4,7,16 As the Down syndrome–critical region on chromosome 21 overlaps with the common region of amplification in iAMP21, coupled with the observation that Down syndrome ALL patients also have a high incidence of gain of chromosome X17 and P2RY8-CRLF2,18 it was speculated that iAMP21 ALL may be related to Down syndrome ALL. If this is so, then the relationship is difficult to discern, as iAMP21 has been seen in only a single Down syndrome ALL patient to date.4 However, other constitutional abnormalities of chromosome 21, in particular the Robertsonian translocation, rob(15;21)(q10;q10)c, are related to iAMP21.4
Mechanism of formation
The complex structure and variability of the iAMP21 chromosome was sufficiently intriguing to warrant detailed genomic study in an attempt to elucidate the mechanism behind its formation. Earlier, it had been shown from extensive FISH mapping that the abnormal chromosome 21 arose through a breakage-fusion-bridge (BFB) mechanism,10 supported by the observation of anaphase bridges involving chromosome 21 in some iAMP21 patients.19 Array-based studies in conjunction with FISH had indicated gross copy-number changes and complex structural rearrangements.7,11 Further analyses pointed to clustered breakpoints within the PDE9A gene in a number of patients involved in complex events around microhomology-mediated end joining as preceding or initiating the BFB cycles.20 These observations raised hope that breakpoints within PDE9A may have been linked to the initiation of the leukemogenic process of iAMP21. However, no further evidence has emerged.
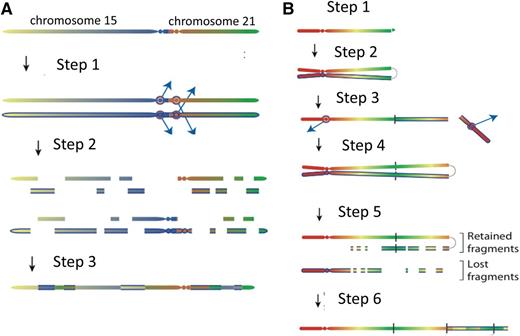
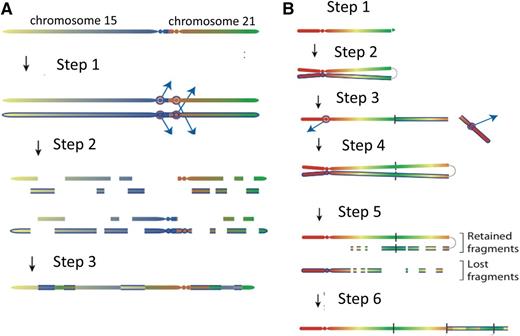
An important observation that illuminated the mechanism came from the fascinating discovery that individuals born with the rare constitutional Robertsonian translocation between chromosomes 15 and 21, rob(15;21)(q10;q10)c, have ∼2700-fold increased risk of developing iAMP21 ALL compared with the general population.21 This is a striking enrichment considering the rarity of this constitutional change. In total, Robertsonian translocations are found in ∼1 in 1000 newborns,22,23 but rob(15;21)c accounts for only 0.5% to 1% of them. The association is remarkably specific, as these individuals appear not to be predisposed to other subtypes of ALL or hematologic malignancies. Intriguingly this strong association with iAMP21 ALL was not seen among the other more frequently occurring Robertsonian translocations. Genomic, cytogenetic, and transcriptional analyses, coupled with next-generation sequencing and novel bioinformatics approaches, were used to identify genomic rearrangements and reconstruct the common principles underlying the temporal evolution of iAMP21 ALL. In the rob(15;21)c cases, amplification was initiated by chromothripsis, a process whereby localized genomic regions are shattered and rearranged in a one-off catastrophic event, simultaneously involving both sister chromatids of the dicentric Robertsonian chromosome21 (Figure 2). This finding implied that this constitutional, structural abnormality was specifically predisposed to leukemia through a novel mechanism, namely a propensity to undergo chromothripsis, likely related to its dicentric nature. One striking common feature of the rob(15;21)c-associated iAMP21 chromosomes was the loss of the chromosome 15 centromere, likely as a result of chromothripsis, providing mitotic stability of the derivative chromosome.
Models for the evolution of the iAMP21 chromosome. (A) The rob(15;21)c is a constitutional dicentric chromosome fusing the short arms of 1 copy each of chromosomes 15 and 21. Usually, 1 centromere is inactive to allow ordered separation of chromatids in mitosis. The model depicts the hypothesis that the 2 centromeres become active and confound attachment of mitotic spindles to the sister kinetochores, such that each chromatid connects to spindles emanating from opposite poles (step 1). During anaphase, this merotelic attachment would lead to lagging of both sister chromatids, rendering them jointly prone to chromothripsis (shattering and then repair) (step 2), leading to a grossly rearranged rob(15;21)c-associated iAMP21 (step 3). (B) Sporadic-iAMP21 formation is initiated by telomere attrition or double-strand breakage (step 1). Following replication, the unprotected chromosome ends fuse (step 2), leading to the formation of anaphase bridges and mitotic chromosome double-strand breakage typical of BFB cycles (step 3). This process is often repeated (step 4). Chromothripsis of the resultant dicentric chromosome is initiated (step 5) in the same manner as described for the rob(15;21)c-associated iAMP21 (step 2 in Figure 1A). The final step (step 6) is repair of the shattered chromosome leading to generation of a stable abnormal chromosome 21.
Models for the evolution of the iAMP21 chromosome. (A) The rob(15;21)c is a constitutional dicentric chromosome fusing the short arms of 1 copy each of chromosomes 15 and 21. Usually, 1 centromere is inactive to allow ordered separation of chromatids in mitosis. The model depicts the hypothesis that the 2 centromeres become active and confound attachment of mitotic spindles to the sister kinetochores, such that each chromatid connects to spindles emanating from opposite poles (step 1). During anaphase, this merotelic attachment would lead to lagging of both sister chromatids, rendering them jointly prone to chromothripsis (shattering and then repair) (step 2), leading to a grossly rearranged rob(15;21)c-associated iAMP21 (step 3). (B) Sporadic-iAMP21 formation is initiated by telomere attrition or double-strand breakage (step 1). Following replication, the unprotected chromosome ends fuse (step 2), leading to the formation of anaphase bridges and mitotic chromosome double-strand breakage typical of BFB cycles (step 3). This process is often repeated (step 4). Chromothripsis of the resultant dicentric chromosome is initiated (step 5) in the same manner as described for the rob(15;21)c-associated iAMP21 (step 2 in Figure 1A). The final step (step 6) is repair of the shattered chromosome leading to generation of a stable abnormal chromosome 21.
Further, it was confirmed that iAMP21 in non-rob(15;21)c patients (sporadic iAMP21) was typically initiated by BFB events, leading to the formation of a dicentric chromosome 21. These chromosomes then become prone to chromothripsis or other rearrangements (Figure 2), as described for rob(15;21)c. Thus, inherent or acquired dicentric chromosomes appear to be the trigger of chromosomal instability. In both rob(15;21)c and sporadic iAMP21, the final stages in the formation of the abnormal chromosome frequently involve large-scale duplications, which take place after chromothripsis. Thus, chromothripsis may be remodeling chromosome 21 in a nonrandom fashion, leading to a stable derivative of chromosome 21 or the rob(15;21)c chromosome with leukemic potential. Chromothripsis is known to occur in other cancer types in a random fashion. This observation of the involvement of chromothripsis in the consistent formation of a specific chromosomal abnormality (iAMP21) is not only the first in ALL but also the first to show that genomic instability in the form of large-scale copy-number changes and other rearrangements can be coordinated in an ordered and sequential fashion. The remaining question is: how is this coordinated process initiated?
Clinical features
iAMP21 patients comprise 2% of childhood BCP-ALL, with roughly equal numbers of males and females, indicating a higher proportion of females than in other ALL subtypes.24,25 Patients are older, with a median age of 9 years (range 2-30 years). The upper age limit remained consistent even when adult ALL series were examined. Approximately 50% are classified as high risk based on age ≥10 years, although they generally have low white cell counts.4
Outcome
A significant finding was that patients with iAMP21 had a poor 5-year EFS rate when treated on standard therapy, compared with other BCP-ALL patients treated on the same protocols.26-28 The relapse rate was high, with a 3-fold increase compared with other BCP-ALL patients, and the temporal pattern of relapse was unusual, appearing constant over time, indicating the occurrence of both early and late relapses. Although the overall survival (OS) rate of 71% was also significantly worse compared with other patients, the large difference between EFS and OS indicated that some patients were being salvaged by postrelapse therapy. Based on these retrospective findings, in the United Kingdom, it was elected to treat iAMP21 patients as very high risk regardless of other risk factors. Thus, on the successor trial, iAMP21 patients were prospectively identified, stratified as high risk, and treated on the most intensive treatment arm. This proved to be successful, with highly statistically significant improvements in 5-year EFS (from 29% to 78%), relapse risk (reduced from 70% to 16%), and OS (from 67% to 89%).24 Similar findings were seen in a pair of contemporary Children’s Oncology Group (COG) trials.25 When children with standard-risk ALL were treated less intensively by COG, those with iAMP21 had a significantly inferior outcome compared with patients without iAMP21. In contrast, children with high-risk ALL were treated more intensively by COG, and there was no significant difference in the outcome of those with or without iAMP21. Concurring data were obtained from a large study from the International Childhood ALL Working Group (Ponte di Legno group), in which iAMP21 patient data were collected from 18 different study groups.4 The same improved response was shown when iAMP21 patients were treated as high risk in these multiple treatment centers, regardless of the backbone chemotherapy regimen given.
Studies on the impact of minimal residual disease (MRD) in these patients have produced conflicting results. The Berlin Frankfurt Munster group found that iAMP21 patients who were MRD positive had an inferior outcome compared with MRD-negative patients,28 whereas the results from the recent COG study suggested that MRD was not of prognostic relevance in this subgroup.25 However, the COG and UK studies concurred in their conclusions that iAMP21 patients should be assigned to the high-risk group and be treated intensively irrespective of MRD. These modifications are now in clinical practice, whereas the Berlin Frankfurt Munster group has not made such changes, because its rare, low-risk MRD iAMP21 patients had only a moderate relapse risk on current therapy.29
The complex chromosomal abnormality, iAMP21, defines a novel cytogenetic subgroup of BCP-ALL with an unusual mechanism behind its formation. At the clinical level, it has now been conclusively demonstrated that treatment of iAMP21 patients as high risk provides a significant improvement in their outcome. It is possible that further genomic investigations into the intriguing structural changes may reveal a number of genes implicated in driving the leukemic process. Potentially, such genes may provide molecular targets for therapy, ultimately reducing the level of toxic treatment currently administered to these patients.
Acknowledgments
This work was supported by Leukaemia and Lymphoma Research, the Wellcome Trust, and the European Research Council.
Authorship
Contribution: C.J.H. wrote the manuscript.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Christine J. Harrison, Leukaemia Research Cytogenetics Group, Northern Institute for Cancer Research, Newcastle University, Level 5, Sir James Spence Institute, Royal Victoria Infirmary, Newcastle-upon-Tyne NE1 4LP, United Kingdom; e-mail: christine.harrison@newcastle.ac.uk.