To the editor:
Granulocyte colony-stimulating factor (G-CSF)–mobilized peripheral blood (MPB) is the preferred source of hematopoietic stem cells (HSCs) and has outcompeted bone marrow for HSC-based therapies.1 Umbilical cord blood (UCB) represents an alternative source of HSCs commonly used in preclinical investigations, but limited cell dose has precluded its wide clinical utility.2 Purification of HSCs to homogeneity from these various sources is essential to better understand their unique biology and identify novel therapeutic strategies targeting these cells. In humans, the CD34+CD38−CD45RA−CD90+ phenotype is now customarily accepted to define a cell population highly enriched in HSCs.3 Recently, Notta et al4 further refined this phenotype with the addition of CD49f, an integrin known to mediate HSC niche interactions.5 Using UCB, they estimated that 1:10 cells within the CD34+CD38−CD45RA−CD90+CD49f+ population had long-term repopulating activity in vivo and could be distinguished from transiently engrafting multipotent progenitors (MPPs) defined as CD34+CD38−CD45RA−CD90−CD49f−. When these highly purified UCB HSCs were further sorted based on high efflux of the mitochondrial dye rhodamine 123 (Rholo),6 an additional twofold HSC enrichment was obtained and, strikingly, a fraction of murine recipients transplanted with single-sorted CD34+CD38−CD45RA−CD90+CD49f+Rholo cells displayed multilineage chimerism. Hence, this report was the first to demonstrate purification of human HSCs at single-cell resolution from UCB. In this study, we investigated whether this panel of 6 markers established in UCB could also mark HSCs in MPB.
Leukapheresis was performed on 14 healthy volunteers following 5 days of G-CSF mobilization. CD34+ cells were enriched from the mononuclear cell (MNC) concentrates. Details of the experimental approach can be found in supplemental Methods (available on the Blood Web site). We first investigated whether markers used to enrich for HSCs and MPPs in UCB could also be used to phenotypically identify these 2 subsets in MPB cells. The average frequency of CD34+ cells within the G-CSF–MPB MNC concentrates was 1 in 100 cells (1.05% ± 0.10%; Figure 1A) with purity ≥98.8%. Using stringent gates (Figure 1B), we found that, on average, 1 in 145 000 MPB MNCs (0.00069% ± 0.00008%) had an HSC phenotype, and 1 in 17 000 MPB MNCs were phenotypically defined as MPPs (0.0059% ± 0.0007%) (Figure 1A). Similar results were obtained in MPB MNCs derived from the phylogenetically related rhesus macaque (supplemental Figure 1A-B) and from 6 healthy subjects mobilized with plerixafor (supplemental Figure 1C-D). Overall, these data indicate a highly consistent pattern of expression of these markers in various sources of MPB cells.
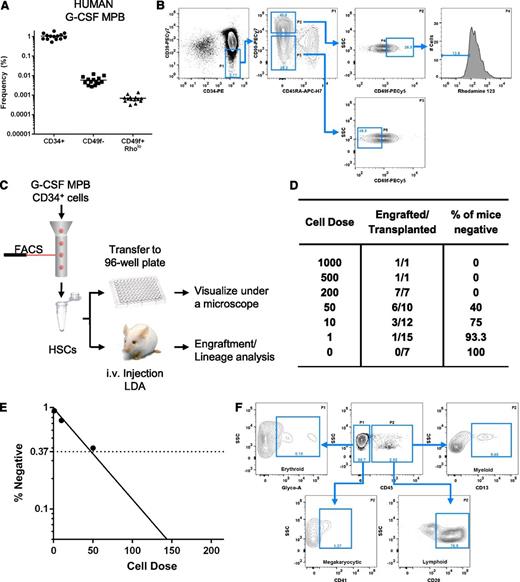
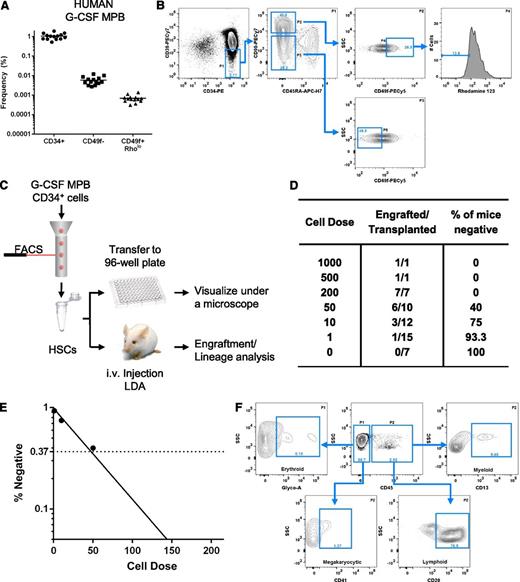
Phenotypic analysis and engraftment potential of human G-CSF–MPB. Phenotypic analysis of G-CSF–MPB MNCs (A-B). (A) Frequency of human CD34+ cells and phenotypically defined MPPs (CD34+CD38−CD45RA−CD90−CD49f−, abbreviated CD49f−) and HSCs (CD34+CD38−CD45RA−CD90+CD49f+Rholo, abbreviated CD49f+Rholo) in G-CSF-MPB MNCs obtained from 14 healthy subjects. (B) Representative flow cytometry analysis for the detection of phenotypically defined MPPs and HSCs in G-CSF-MPB MNCs; the frequency of each subpopulation is based on the parent gate shown on the top right of each plot. Using a limiting dilution transplantation approach, the frequency of HSC within the CD34+CD38−CD45RA−CD90+CD49f+Rholo subfraction was determined (C-E). (C) Schematic of the experimental design. (D) Table outlining the number of mice transplanted at each dose, the number of mice that engrafted, and the percentage of mice that failed to engraft at each respective dose. (E) Semilogarithmic plot of the frequency of long-term repopulating cells within the CD34+CD38−CD45RA−CD90+CD49f+Rholo subfraction measured by extreme limiting dilution analysis. (D) Representative flow cytometry analysis from a single NSG mouse recipient transplanted with 200 CD34+CD38−CD45RA−CD90+CD49f+Rholo cells for human cell engraftment (CD45) and lineage contribution. Analysis is representative of 3 independent experiments (n = 7 mice).
Phenotypic analysis and engraftment potential of human G-CSF–MPB. Phenotypic analysis of G-CSF–MPB MNCs (A-B). (A) Frequency of human CD34+ cells and phenotypically defined MPPs (CD34+CD38−CD45RA−CD90−CD49f−, abbreviated CD49f−) and HSCs (CD34+CD38−CD45RA−CD90+CD49f+Rholo, abbreviated CD49f+Rholo) in G-CSF-MPB MNCs obtained from 14 healthy subjects. (B) Representative flow cytometry analysis for the detection of phenotypically defined MPPs and HSCs in G-CSF-MPB MNCs; the frequency of each subpopulation is based on the parent gate shown on the top right of each plot. Using a limiting dilution transplantation approach, the frequency of HSC within the CD34+CD38−CD45RA−CD90+CD49f+Rholo subfraction was determined (C-E). (C) Schematic of the experimental design. (D) Table outlining the number of mice transplanted at each dose, the number of mice that engrafted, and the percentage of mice that failed to engraft at each respective dose. (E) Semilogarithmic plot of the frequency of long-term repopulating cells within the CD34+CD38−CD45RA−CD90+CD49f+Rholo subfraction measured by extreme limiting dilution analysis. (D) Representative flow cytometry analysis from a single NSG mouse recipient transplanted with 200 CD34+CD38−CD45RA−CD90+CD49f+Rholo cells for human cell engraftment (CD45) and lineage contribution. Analysis is representative of 3 independent experiments (n = 7 mice).
We next performed limiting dilution analysis7 to measure the frequency of HSCs within the CD34+CD38−CD45RA−CD90+CD49f+Rholo population using G-CSF–MPB cells (Figure 1C). A total of 53 NSG mice were transplanted by intravenous route and engraftment was defined as ≥0.1% human CD45+ cells. Limiting dilution analysis indicates that 2.3% (1 in 43 cells; 95% confidence interval, 1 in 23.3 to 1 in 78.4 cells) of the G-CSF–MPB CD34+CD38−CD45RA−CD90+CD49f+Rholo fraction has long-term repopulating activity in NSG mice (Figure 1D-E). One of 15 animals transplanted with a single cell displayed robust engraftment (9.2% human CD45+ cells; data not shown). Additionally, all major hematopoietic lineages could be detected in the bone marrow of engrafted mice (n = 7), including cells of the erythroid (2.9% ± 2.5%), myeloid (8.2% ± 4.2%), megakaryocytic (6.6% ± 2.7%), and B-lymphoid (75.4% ± 4.5%) lineages (Figure 1F).
In this study, we provide evidence that CD49f can be used in combination with common HSC markers to enrich cells with long-term engraftment potential from G-CSF-MPB samples. Detection of engraftment in one animal transplanted with a single cell indicates that human HSCs derived from G-CSF-MPB samples express CD49f. Because 1 in 145 000 G-CSF–MPB MNCs are CD34+CD38−CD45RA−CD90+CD49f+Rholo (Figure 1A) and 2.3% of this fraction has repopulating activity (Figure 1D-E), the overall HSC frequency in G-CSF–MPB MNCs is estimated at 1 in 6.2 × 106 cells, consistent with a previously reported estimate using this model.8 These data contribute to the body of evidence indicating a five- to 10-fold lower HSC frequency in MPB compared with UCB sources.8 However, because the total number of CD34+ cells obtained after G-CSF mobilization of a healthy subject is on average 100- to 200-fold superior to CD34+ cell numbers obtained from a single UCB unit, MPB samples represent a more practical source of HSCs and MPPs for isolation of sufficient numbers of cells required for both clinical applications and fundamental investigations of the molecular determinants of these cell populations.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank David Stroncek and the National Institutes of Health Department of Transfusion Medicine and Cell Processing Section staff for apheresis, selection, and cryopreservation of human CD34+ cells; Roumiana Nenkova-Dimtcheva and the Hematology Branch outpatient clinic nursing staff for providing G-CSF administration teaching to healthy subjects; and Crystal Thomas and the mouse core facility staff, as well as Robert E. Donahue and the nonhuman primate core facility staff, for excellent animal care. This work was supported by the intramural research program of the National Heart, Lung and Blood Institute of the National Institutes of Health.
Contribution: H.D.H., T.B., and A.L. designed and performed the experimental procedures, analyzed the data, and wrote and edited the manuscript; H.C., A.C., P.S.C., and J.-F.F. performed some of the flow stains and analyses; K.K. sorted the cell populations; R.W.C. provided the AMD3100-mobilized samples; and C.E.D. edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Andre Larochelle, National Heart, Lung and Blood Institute, National Institutes of Health, Bethesda, MD 20892; e-mail: larochea@nhlbi.nih.gov.
References
Author notes
H.D.H. and T.B. contributed equally to this study.