Key Points
Wnt/β-catenin signaling increases ETO and Runx1 transcription in human hematopoietic progenitors.
Wnt/β-catenin signaling enhances spatial proximity of ETO and RUNX1 genes and induces the generation of a recurrent translocation event.
Abstract
Chromosomal translocations are frequently associated with a wide variety of cancers, particularly hematologic malignancies. A recurrent chromosomal abnormality in acute myeloid leukemia is the reciprocal translocation t(8;21) that fuses RUNX1 and ETO genes. We report here that Wnt/β-catenin signaling increases the expression of ETO and RUNX1 genes in human hematopoietic progenitors. We found that β-catenin is rapidly recruited into RNA polymerase II transcription factories (RNAPII-Ser5) and that ETO and RUNX1 genes are brought into close spatial proximity upon Wnt3a induction. Notably, long-term treatment of cells with Wnt3a induces the generation a frequent RUNX1-ETO translocation event. Thus, Wnt/β-catenin signaling induces transcription and translocation of RUNX1 and ETO fusion gene partners, opening a novel window to understand the onset/development of leukemia.
Introduction
One of the most common chromosomal abnormalities in acute myeloid leukemia (AML) is the reciprocal translocation t(8;21)(q22;q22), which involves the RUNX1 (AML1) and the ETO (RUNX1T1) genes and produces a Runx1-ETO fusion transcript that inhibits Runx1-dependent transcriptional regulation.1,2 Although RUNX1-ETO translocations could act as initiating events in hematopoietic stem cells (HSCs), after which leukemia clonally evolves through the acquisition of secondary mutations,3,4 the molecular mechanism and cellular signals that drive the generation of the t(8;21) translocation remain to be elucidated.
Although much has been learned in recent years about the onset or development of AML from studies examining Runx1 expression or function,5-8 we are still far from a complete understanding of the cellular mechanisms controlling the transcription of its translocation partner, ETO.9-11 Considering that Wnt/β-catenin plays essential roles during the proliferation or differentiation of HSCs and that reactivation of β-catenin signaling is important for self-renewal of leukemia stem cells,12-14 we studied whether Wnt/β-catenin signaling was involved in ETO expression and RUNX1-ETO fusion.
Methods
Human CD34+ cells and cell lines
CD34+ cells were isolated from umbilical cord blood samples after normal full-term delivery from the Maternity Service at Complejo Asistencial Barros Luco (Santiago, Chile). Informed consent was obtained in accordance with the Declaration of Helsinki and the ethics committee of the Universidad Andres Bello. KG1, HEK293, and L cells secreting Wnt3a were obtained from ATCC (Rockville, MD).
Cell culture conditions, Wnt/β-catenin signaling activity, semiquantitative and quantitative polymerase chain reaction, ETO promoter activity, plasmids and site-directed mutagenesis, β-catenin and RNAPII immunofluorescence, RNA and DNA fluorescent in situ hybridization (FISH) experiments, image acquisition and processing, and statistical analyses are described in supplemental Methods (available on the Blood Web site).
Results and discussion
According to the current assembly of the human genome (GRCh38), the ETO gene consists of 17 exons distributed over 148 kb (Figure 1A), and its expression generates multiple messenger RNA (mRNA) isoforms by alternative splicing or the use of different promoters.9-11 We searched for core TCF/LEF-binding elements (TBE: CTTTG) upstream and downstream of the proposed transcriptional start sites of the human gene and found multiple TBE sites in these genomic regions. We designed primers to detect common alternative mRNA transcripts (HGNC: ETO-001, 002, 004, and 009; supplemental Table 1). Although endogenous ETO mRNA levels were readily detected in isolated CD34+ cells under control conditions,6 we observed that a differential enhancement in the transcription of ETO-004 variant occurred in CD34+ cells after short-term treatment (4 hours) with purified Wnt3a (Figure 1B). Similarly, whereas endogenous levels of ETO mRNA were barely detected in the KG1 cells15 under control conditions, a clear increase in the expression of ETO-004 variant occurred in these cells upon Wnt3a treatment. ETO variants were also detected in HEK293 cells under control or Wnt3a conditions, suggesting that this embryonic cell line could be a useful model to study alternative ETO expression.
Wnt/β-catenin signaling modulates ETO transcriptional activity. (A) Genomic context of the human ETO gene on chromosome 8. Selected ETO mRNA variants (001: ENST00000523629; 002: ENST00000396218; 004: ENST00000422361; and 009: ENST00000360348) (top). Exons are depicted as bars numbered according to NCBI RefSeq: NC_000008.11. Numbers within square brackets indicate numbering as it has been reported.9 Green boxes (P1-P8) represent predicted genomic regions containing putative promoters.11 t(8,21): Chromosomal translocation sites RUNX1/ETO. The location of predicted TCF/LEF-binding elements (TBE; open circles) in the promoter region of variants 002, 004, and 009 (bottom). (B) Detection of alternative ETO mRNA variants 001, 002, 004, and 009 in CD34+ (top), KG1 (middle), and HEK293 cells (bottom) under control conditions or in the same cells treated with purified Wnt3a (200 ng/mL) for 4 hours. (C) Quantitative determination of mRNA levels for ETO-004 and ETO total (using primers designed in conserved exons 8 and 9 or determined by TaqMan assays; left and middle, respectively), as well as for Runx1 and Wnt/β-catenin target genes Axin2 and Myc (right). Cells were incubated under control conditions or stimulated with 200 ng/mL purified Wnt3a for 4 hours. Quantitative polymerase chain reaction results were normalized using GAPDH mRNA levels as a reference gene and are expressed as relative expression. Each figure corresponds to at least 5 independent experiments (with replicates). Ctrl, control conditions. Statistical significance was determined by a Mann-Whitney U test. (D) Dose-dependent effects of constitutively active β-catenin S33Y on the transcriptional activity of ETO-luciferase promoter constructs in HEK293 cells (24 hours). SuperTOPFLASH (STF) was measured as positive Wnt/β-catenin signaling readout. Each figure corresponds to at least 3 independent experiments. Statistical significance was determined by an ANOVA test (**P < .01). (E) Effect of β-catenin S33Y and increasing doses of dominant-negative ΔTCF4 on the promoter activity of pETO-004 deletion constructs, determined as in panel D. RLU, relative luciferase units.
Wnt/β-catenin signaling modulates ETO transcriptional activity. (A) Genomic context of the human ETO gene on chromosome 8. Selected ETO mRNA variants (001: ENST00000523629; 002: ENST00000396218; 004: ENST00000422361; and 009: ENST00000360348) (top). Exons are depicted as bars numbered according to NCBI RefSeq: NC_000008.11. Numbers within square brackets indicate numbering as it has been reported.9 Green boxes (P1-P8) represent predicted genomic regions containing putative promoters.11 t(8,21): Chromosomal translocation sites RUNX1/ETO. The location of predicted TCF/LEF-binding elements (TBE; open circles) in the promoter region of variants 002, 004, and 009 (bottom). (B) Detection of alternative ETO mRNA variants 001, 002, 004, and 009 in CD34+ (top), KG1 (middle), and HEK293 cells (bottom) under control conditions or in the same cells treated with purified Wnt3a (200 ng/mL) for 4 hours. (C) Quantitative determination of mRNA levels for ETO-004 and ETO total (using primers designed in conserved exons 8 and 9 or determined by TaqMan assays; left and middle, respectively), as well as for Runx1 and Wnt/β-catenin target genes Axin2 and Myc (right). Cells were incubated under control conditions or stimulated with 200 ng/mL purified Wnt3a for 4 hours. Quantitative polymerase chain reaction results were normalized using GAPDH mRNA levels as a reference gene and are expressed as relative expression. Each figure corresponds to at least 5 independent experiments (with replicates). Ctrl, control conditions. Statistical significance was determined by a Mann-Whitney U test. (D) Dose-dependent effects of constitutively active β-catenin S33Y on the transcriptional activity of ETO-luciferase promoter constructs in HEK293 cells (24 hours). SuperTOPFLASH (STF) was measured as positive Wnt/β-catenin signaling readout. Each figure corresponds to at least 3 independent experiments. Statistical significance was determined by an ANOVA test (**P < .01). (E) Effect of β-catenin S33Y and increasing doses of dominant-negative ΔTCF4 on the promoter activity of pETO-004 deletion constructs, determined as in panel D. RLU, relative luciferase units.
Quantitative determination of mRNA levels in CD34+ cells showed that Wnt/β-catenin induced a rapid (4-hour) and significant increase in ETO-004 variant (3.7-fold, P = .0028, n = 6) or total ETO mRNA levels (3.6-fold, P = .0028, n = 6; Figure 1C). Wnt3a effects on total ETO levels were further confirmed by TaqMan analyses (2.9-fold, P = .0011, n = 7; Figure 1C and supplemental Figure 1). Finally, the effect of the signaling cascade on ETO expression was paralleled by enhanced transcription of Runx116,17 (2.6-fold, P = .0075, n = 5) and classical target genes Axin2 and Myc (3.5- and 2.8-fold, respectively; P = .0075, n = 5).18,19
Putative promoters for ETO-002 and -004 variants are located downstream of common RUNX1-ETO translocation hotspots (Figure 1A). To study if these promoters contained functional TBE sites, we PCR-cloned 890 and 2459 nucleotides of the 5′ upstream/exon 1 sequences of ETO-002 and -004, containing 6 and 7 potential TBE sites, respectively (Figure 1D). We also analyzed 1535 nucleotides with 5 TBE sites upstream of the transcriptional start sites described for variant ETO-009.10 We observed a significant increase in the promoter activity of pETO-004 (2.5 kb) in HEK293 cells cotransfected for 24 hours with increasing concentrations of constitutive active β-catenin (S33Y) (Figure 1D). Although basal transcription was observed for pETO-002 and pETO-009 promoters, no significant effects were found in cells cotransfected with these constructs and β-catenin S33Y (24 hours). Then, we serially deleted potential TBE sites in pETO-004 (pETO-004-0.9 and pETO-004-04, respectively) and observed that these constructs responded accordingly to β-catenin stabilization or its inhibition by a dominant-negative ΔTCF4 construct (Figure 1E). Next, because pETO-004-0.4 contains a single TBE site (−457 nucleotides upstream of the ATG of ETO-004 variant), we mutated the core consensus sequence (CTTTG → CCTCG) and found that the activity of the mutant promoter was decreased to basal levels (supplemental Figure 2). Collectively, our findings indicate that Wnt/β-catenin signaling rapidly induces ETO-004 variant and that this effect is probably accomplished through ETO-004 TBE site I.
Translocation-prone genes are preferentially recruited into transcription factories with active RNA polymerase II (RNAPII-Ser5)20,21 and need to be positioned in close spatial proximity relative to each other prior to translocation.22,23 Because β-catenin participates in recruiting basal transcription components at the promoter of its target genes, we studied if activation of the cascade induced β-catenin relocalization into these transcriptional structures. We found that transcription factories labeled with active RNAPII-Ser5 were clearly distinguished in resting or Wnt3a-treated CD34+ cells (4 hours) (Figure 2A-B). Under control conditions, the number and diameter of RNAPII speckles within the nucleus were in agreement with previous observations22 (32.1 ± 1.13 and 130.7 ± 3.5 nm, respectively; supplemental Figure 3A). Remarkably, Wnt3a induced a significant increase in the number of endogenous β-catenin speckles within the nucleus of CD34+ cells (control: 1.5 ± 0.3; Wnt3a: 19.5 ± 2.7; P < .001; supplemental Figure 3A), some of which clearly colocalized with active RNAPII foci (supplemental Figure 3B-C). Similar effects of β-catenin colocalization with RNAPII-Ser5 were observed in Wnt3a-treated KG1 cells (supplemental Figure 4).
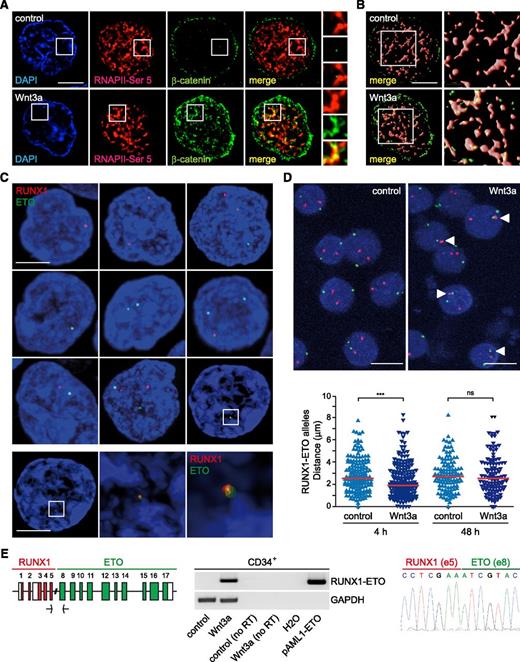
Wnt3a induces spatial proximity and translocation of RUNX1 and ETO in human hematopoietic progenitors. (A) Localization of β-catenin into active RNAPII transcription factories. Immunodetection of β-catenin and active RNAPII-Ser5P through confocal microscopy in CD34+ cells incubated under control conditions or in the presence of purified Wnt3a (200 ng/mL; 4 hours). White boxes indicate selected regions of interest that are shown at higher magnification at the right panel. Scale bar represents 5 μm. (B) Surface-rendered CD34+ cells (as depicted in panel A) and zoom in (4×; white boxes) showing colocalization of β-catenin and RNAPII-Ser5P in Wnt3a-treated cells. (C) Representative confocal images of RNA-FISH experiments in CD34+ cells upon 4-hour treatment with 200 ng/mL purified Wnt3a (number of cells and other parameters are given in supplemental Table 2). RUNX1 alleles are shown in red and ETO alleles in green. Scale bar represents 4 μm. Selected region of interest (white box) is shown at higher magnification below as a maximum intensity projection reconstruction for 4,6 diamidino-2-phenylindole (DAPI) and signal rendering for RUNX1 (red) and ETO (green). Scale bar represents 4 μm. Zoom in (4× and 8×; middle and right, respectively) corresponds to region of interest defined above. (D) Representative confocal images of DNA-FISH experiments in control and Wnt3a-treated CD34+ cells (4 hours) (top). White arrowheads in Wnt3a-treated cells indicate RUNX1 (red) and ETO (green) alleles with a distance of less than 1 μm. Scale bar represents 10 μm. Scatterplot showing shortest distances between RUNX1 and ETO alleles in control and Wnt3a-treated cells for 4 and 48 hours (bottom) (number of cells and other parameters are given in supplemental Table 3). The median in each group is depicted as a red line. Statistical significance was determined by a Mann-Whitney U test (***P < .0001; NS, nonsignificant). (E) Wnt/β-catenin signaling induces a common RUNX1-ETO translocation event. Diagram representing a recurrent RUNX1-ETO translocation event (left). Detection of RUNX1-ETO fusion gene and GAPDH products in CD34+ cells following long-term treatment with Wnt3a for 48 hours (middle). Chromatogram of the band shown for CD34+ cells treated with Wnt3a (48 hours) (right). no RT, no reverse transcription; pAML1-ETO, amplification of RUNX1-ETO from chimeric plasmid pCMV5-AML1-ETO.
Wnt3a induces spatial proximity and translocation of RUNX1 and ETO in human hematopoietic progenitors. (A) Localization of β-catenin into active RNAPII transcription factories. Immunodetection of β-catenin and active RNAPII-Ser5P through confocal microscopy in CD34+ cells incubated under control conditions or in the presence of purified Wnt3a (200 ng/mL; 4 hours). White boxes indicate selected regions of interest that are shown at higher magnification at the right panel. Scale bar represents 5 μm. (B) Surface-rendered CD34+ cells (as depicted in panel A) and zoom in (4×; white boxes) showing colocalization of β-catenin and RNAPII-Ser5P in Wnt3a-treated cells. (C) Representative confocal images of RNA-FISH experiments in CD34+ cells upon 4-hour treatment with 200 ng/mL purified Wnt3a (number of cells and other parameters are given in supplemental Table 2). RUNX1 alleles are shown in red and ETO alleles in green. Scale bar represents 4 μm. Selected region of interest (white box) is shown at higher magnification below as a maximum intensity projection reconstruction for 4,6 diamidino-2-phenylindole (DAPI) and signal rendering for RUNX1 (red) and ETO (green). Scale bar represents 4 μm. Zoom in (4× and 8×; middle and right, respectively) corresponds to region of interest defined above. (D) Representative confocal images of DNA-FISH experiments in control and Wnt3a-treated CD34+ cells (4 hours) (top). White arrowheads in Wnt3a-treated cells indicate RUNX1 (red) and ETO (green) alleles with a distance of less than 1 μm. Scale bar represents 10 μm. Scatterplot showing shortest distances between RUNX1 and ETO alleles in control and Wnt3a-treated cells for 4 and 48 hours (bottom) (number of cells and other parameters are given in supplemental Table 3). The median in each group is depicted as a red line. Statistical significance was determined by a Mann-Whitney U test (***P < .0001; NS, nonsignificant). (E) Wnt/β-catenin signaling induces a common RUNX1-ETO translocation event. Diagram representing a recurrent RUNX1-ETO translocation event (left). Detection of RUNX1-ETO fusion gene and GAPDH products in CD34+ cells following long-term treatment with Wnt3a for 48 hours (middle). Chromatogram of the band shown for CD34+ cells treated with Wnt3a (48 hours) (right). no RT, no reverse transcription; pAML1-ETO, amplification of RUNX1-ETO from chimeric plasmid pCMV5-AML1-ETO.
Our data indicate that β-catenin rapidly associates with RNAPII transcription factories in response to Wnt/β-catenin activation, further implying that expression of ETO and RUNX1 translocation partners could occur in these chromosomal structures. To address this idea, we performed RNA-FISH experiments in CD34+ cells using fluorescent probes targeting nascent sequences within ETO and RUNX1 genes (introns 11 and 4, respectively) (Figure 2C). We noted that the total number of ETO and RUNX1 alleles expressed in control CD34+ cells increased from 25 to 54 after 4-hour Wnt3a-treatment (supplemental Figure 5 and supplemental Table 2). Wnt/β-catenin induction was similarly observed for both genes (ETO alleles: from 9 to 21; RUNX1 alleles: from 16 to 33) and turned on the expression of a second allele in a greater number of cells. We determined the distance between RUNX1 and ETO alleles being simultaneously transcribed and observed that from a total of 105 Wnt3a-treated cells, 7 cells had distances <300 nm between alleles (there were no events in the control setting that showed a proximity <300 nm) and that the RNA signals in 3 of these CD34+ cells localized into the same chromosomal foci following activation of the cascade (Figure 2C and supplemental Figure 5). Altogether, these observations confirm that Wnt/β-catenin enhances ETO and Runx1 expression in hematopoietic precursors and indicate that transcription of these genes is likely occurring into specialized nuclear factories. To further examine the hypothesis that Wnt signaling facilitates genomic proximity, we performed DNA-FISH experiments in CD34+ cells and observed that RUNX1 and ETO genes are indeed brought up in close proximity (<1 μm) after short-term Wnt3a treatment (4 hours) (23% vs 10%; Wnt3a vs control conditions, respectively; Figure 2D and supplemental Table 3). No significant differences in proximity were found between RUNX1 and ETO at 48 hours, suggesting that an early transcriptional program commanded by the signaling cascade is involved in chromosome spatial organization and gene regulation.
Recent experiments indicate that β-catenin promotes genomic instability and T-cell transformation in mice by compromising DNA repair and enhancing illegitimate recombination.24 Because most RUNX1-ETO translocations occur within RUNX1 intron 5 and ETO intron 7 we designed primers in RUNX1 exon 5 and ETO exon 8 to detect the product of the fusion of these genes. Notably, in CD34+ cells incubated with Wnt3a for 48 hours, we observed amplification of the expected RUNX1-ETO chimeric product (Figure 2E and supplemental Figure 6). Finally, Wnt/β-catenin effects were further detected in HEK293 cells transfected for 48 hours with increasing doses of β-catenin S33Y (supplemental Figure 7). Our data thus indicate that sustained transcriptional activation induced by the signaling cascade represents an early event that triggers the translocation of RUNX1 and ETO genes.
Deregulation of Wnt/β-catenin signaling as a causative factor in leukemogenesis has become increasingly apparent.12-14 Here, we studied the transcriptional program directed by Wnt/β-catenin signaling in CD34+ and KG1 cells because they resemble normal human HSCs15 and it has been suggested that immature cells, rather than committed progenitors, are the targets for leukemic transformation.25 We observed that gain of function of Wnt/β-catenin signaling enhances transcription of ETO and RUNX1 translocation partners in genome organization centers within the nucleus, thereby increasing the potential for a chromosomal translocation event. To understand the role of Wnt/β-catenin signaling in other recurrent translocations remains a goal of further investigations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to the Maternity Service at Complejo Asistencial Barros Luco. The authors thank Dr Martín Montecino for sharing the pCMV5-AML1-ETO plasmid and Flavia Román and Dr Miguel Avila for technical assistance or preliminary work.
This work was supported by Comisión Nacional de Investigación Científica y Tecnológica (CONICYT) grants Anillo ACT-1119 (G.V.D.F. and A.L.), FONDECYT Postdoctorado 3130509 (G.D.U.) and 3150612 (M.F.V.), Fondo Nacional de Desarrollo Científico y Tecnológico (FONDECYT) Regular 1140353 (G.V.D.F.) and 1130697 (S.E.G.), Chilean Copper Commission (COCHILCO)-FONDECYT 1100995 and IMII P09-016-F (A.E.), and FONDAP-CRG 15090007 (G.V.D.F.). R.T.M. is supported as an Investigator of the Howard Hughes Medical Institute.
Authorship
Contribution: G.V.D.F. and G.D.U. designed the study; G.D.U., M.F.V., M.A.M., P.L., and D.N. performed experiments, analysis, and interpretation of the data; S.E.G., A.A.E., R.T.M., and A.L. helped in drafting the manuscript and contributed cell or molecular tools; and G.D.U., M.F.V., M.A.M., and G.V.D.F. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giancarlo V. De Ferrari, Center for Biomedical Research, FONDAP Center for Genome Regulation, Faculty of Biological Sciences, Universidad Andrés Bello, República 239, Santiago, Chile; e-mail: gdeferrari@unab.cl.
References
Author notes
G.D.U. and M.F.V. contributed equally to this study.