Abstract
SRSF2 is mutated at proline 95 (P95) in approximately 45% of patients with CMML. The consequence of SRSF2 P95H mutations on splicing has been associated with change in target RNA motif preference compared to wild type that favors CCNG over GGNG resulting in aberrant splicing abnormalities. Although this has led to several downstream mis-spliced candidates that may contribute to SRSF2 leukemic pathogenesis, the biologic consequences of SRSF2 mutation have not been fully elucidated and its effect on non-splicing pathways has yet to be explored.
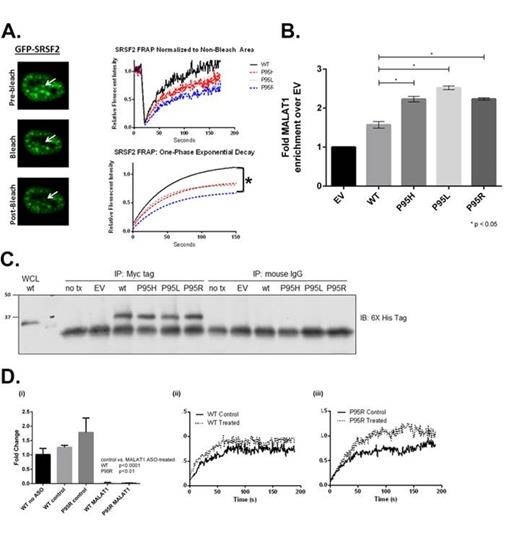
SRSF2 has two major functions within the nucleus: (1) bind to cis elements on pre-mRNA transcripts that functionally redefines putative exon-intron boundaries and (2) interact with other members of the spliceosome complex at nuclear suborganelles known as speckles. To explore the effects of SRSF2 P95 mutation on nuclear speckle dynamics, we transfected HeLa cells with GFP-SRSF2 wildtype (WT) and mutant (MT) constructs and performed Fluorescent Recovery After Photo-bleaching (FRAP) to determine the mutant specific kinetics of SRSF2 molecules localized to the nuclear speckle. Using this approach, we confirmed that SRSF2 WTs are characterized by complete photo-recovery with a half-life of approximately 35 (30-42) seconds as previously reported. However, when we performed FRAP of GFP-SRSF2 MTs a statistically significant difference in photo-recovery was observed compared to WT cells (Fig. 1a). The difference between baseline fluorescence and maximal recovered fluorescence in SRSF2 MT cells, or the immobile fraction, suggests that trapped bleached SRSF2 molecules are sterically inhibiting the diffusion of unbleached SRSF2 molecules.
We reasoned that the observed differences in RNA binding capacity and nuclear speckle dynamics could be related. This rationale identified MALAT1 as a link connecting both increased RNA binding and nuclear speckle trafficking abnormalities because it is a Long Non-Coding (lnc) RNA that localizes to and has a critical role in SR protein recruitment and retention to the nuclear speckle. MALAT1, which is highly enriched for CCNG motifs, has also been demonstrated to bind to SRSF2 directly and has been implicated in a wide spectrum of carcinomas making it an attractive intermediary to study in our system.
To determine whether SRSF2 MTs have increased MALAT1 binding capacity compared to SRSF2 WT, we performed RNA IP of myc-his-SRSF2 WT and MT transfected HeLa cells. Using this approach, we demonstrated a 60% enrichment of MALAT1 in SRSF2 WT transfected cells and a 120-150% enrichment of MALAT1 in SRSF2 MT cells compared to the empty vector control (Fig. 1b-c). We next performed the above FRAP experiment in the context of MALAT1 depletion and demonstrated that observed immobile fraction seen in SRSF2 MT cells is rescued (Fig. 1d).
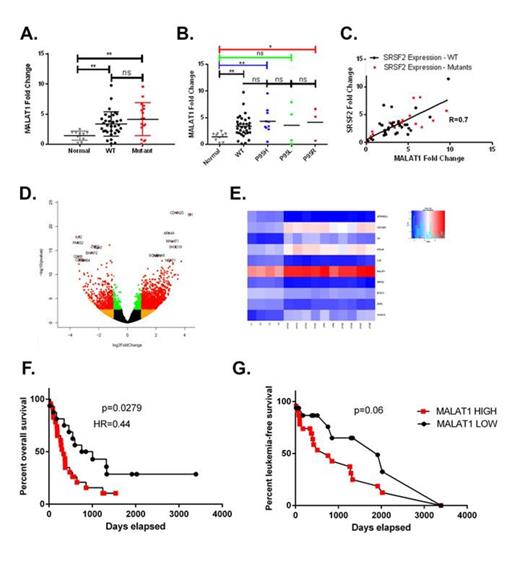
To determine the clinical relevance of MALAT1 in CMML we profiled a cohort of 54 CMML cases for MALAT1 expression by PCR. As shown in Figure 2a-b and d-e, MALAT1 expression is statistically elevated in CMML BMNCs and is among the highest differentially expressed transcripts in CMML monocytes (CD14+). We additionally characterized SRSF2 expression in our CMML BMNCs and CD14+ cohort and identified a linear correlation (Pearson R=0.7 p<0.05) between SRSF2 expression and MALAT1 expression (Figure 2c). Further, CMML patients with high MALAT1 expression in the BMNC compartment had inferior overall and leukemia-free survival (Fig. 2f-g).
Given its relevance in solid tumors and its over-expression in CMML, we explored the consequence of MALAT1 depletion on human monocytic leukemic cells. We first performed ultra-deep RNA sequencing of the THP-1 cell line treated with two MALAT-1 depleting ASOs or matched chemistry specific controls. IPA analysis of differentially expressed transcripts identified a c-Myc gene signature that was validated by western blot analysis demonstrating reduction of c-Myc in THP-1 MALAT1-depleted cells. Colony formation assays using the THP-1 cell line demonstrated that MALAT-1 depletion resulted in decreased colony-forming capacity (CFC). MALAT-1 ASOs were also capable of depleting MALAT-1 in primary CMML BMNCs and were associated with a decreased CFC suggesting that MALAT-1 is a potential therapeutic target. Taken together, our data identifies SRSF2 P95 mutation nuclear trafficking abnormalities and identifies a novel clinically relevant lncRNA in CMML.
Komrokji:Celgene: Consultancy, Research Funding; Incite: Consultancy; Novartis: Speakers Bureau; GSK: Research Funding. List:Celgene Corporation: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.