Abstract
Introduction
JMML is a rare leukemia characterized by aberrant myeloid proliferation and hypersensitivity to GM-CSF. Mutually exclusive mutations in PTPN11, N/K-RAS, CBL, or NF1 are found in ~90% of patients. These mutations cause the disease at least in part by activating STAT5 through phosphorylation and promoting cell growth. MicroRNAs (miRs) are small (~22 nucleotides) noncoding RNAs, regulating gene expression and often deregulated in leukemia. We investigated whether miRs are deregulated in JMML and whether miRs target critical proteins involved in the disease pathogenesis.
Patients and Methods
MiRs expression profile of 40 bone marrow (BM) samples from untreated JMML patients and 8 BM samples from healthy controls was performed using the Ncounter Human v2 miRNA Expression Assay (nanostring). MiR150-5p and STAT5b mRNA expression were assessed using quantitative (q)RT-PCR. Selected miRs and target expression were measured in BM and spleen from a JMML (PTPN11) murine model (D61Y mutation). We overexpressed miR-150-5p in AML cell lines (K562, OCI-AML-3, KG1a) using a miR-150-5p precursor plasmid and STAT5b mRNA was assessed by qRT-PCR. STAT5 and phospho-STAT5 protein levels were assessed by Western Blot in miR-150-5p transfected AML cell lines, human JMML and mice BM cells. STAT5b 3'UTR sequence was cloned in a Firefly/Renilla Luc construct and co-transfected with miR-150-5p miRNA mimic into 293T cell line.
Results
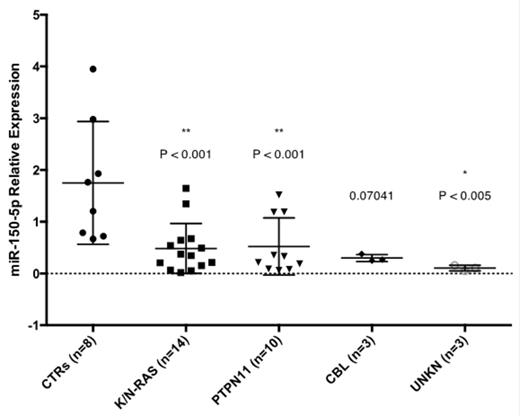
We found that 25 miRs were differentially expressed in whole BM cells from JMML patients respect to healthy controls (Table 1). MiR-150-5p was the most downregulated miR in JMML. We focused on miR-150-5p, since it has been described to be downregulated in AML cases and is predicted to target STAT5b, a critical gene in JMML biology. We validated that miR-150-5p was down-regulated in JMML cases respect to controls performing qRT-PCR on 38 BM samples from JMML patients. Likewise, miR-150-5p was downregulated in BM and spleen samples from PTPN11 mutated mice respect to controls (0.35 and 0.27 Average Fold Change decrease respectively). STAT5 protein, a predicted target for miR-150-5p, was highly expressed in the JMML patients and mice BM samples respect to their controls (4.3 and 1.3 Average Fold Change increase respectively). MiR-150-5p overexpression in K562, OCI-AML-3 and KG1a cell lines led to decrease of STAT5 protein levels and phosphorilation at 48 hours. Direct interaction of STAT5b 3'UTR with miR-150-5p was demonstrated by luciferase assay (~50% Luc activity inhibition, P<0.001). Last, overexpression of miR-150-5p in primary JMML samples using a GFP tagged lentivirus significantly decreased cell growth respect to controls (empty vector) in response to GM-CSF (P<0.01).
Discussion
We showed that miR-150-5p is downregulated in JMML samples and in JMML animal models. Functionally, miR-150-5p directly inhibits the translation of STAT5b mRNA resulting also in a decrease of phosphorylation of STAT5 total protein. These findings identify an alternative mechanism that supports STAT5 deregulation in JMML and that could be therapeutically targeted.
Deregulated miRNAs in JMML
| Gene . | Accession # . | P value . | Fold-Change (Log2) . |
|---|---|---|---|
| miR-150-5p | MIMAT0000451 | 0,001 | -2,382 |
| let-7g-5p | MIMAT0000414 | 0,002 | -1,660 |
| miR-1260a | MIMAT0005911 | 0,009 | -1,592 |
| let-7a-5p | MIMAT0000062 | 0,010 | -1,574 |
| miR-4454 | MIMAT0018976 | 0,021 | -1,399 |
| miR-148a-3p | MIMAT0000243 | 0,030 | -1,211 |
| miR-146b-5p | MIMAT0002809 | 0,009 | -1,084 |
| miR-342-3p | MIMAT0000753 | 0,010 | -1,075 |
| let-7f-5p | MIMAT0000067 | 0,021 | -1,022 |
| miR-26a-5p | MIMAT0000082 | 0,034 | -1,008 |
| let-7d-5p | MIMAT0000065 | 0,038 | -1,005 |
| miR-30b-5p | MIMAT0000420 | 0,019 | -0,972 |
| miR-29b-3p | MIMAT0000100 | 0,044 | -0,956 |
| miR-29a-3p | MIMAT0000086 | 0,024 | -0,761 |
| miR-338-3p | MIMAT0000763 | 0,042 | 0,654 |
| miR-23a-3p | MIMAT0000078 | 0,040 | 0,689 |
| miR-222-3p | MIMAT0000279 | 0,018 | 0,756 |
| miR-548ai | MIMAT0018989 | 0,009 | 0,993 |
| miR-494 | MIMAT0002816 | 0,023 | 1,021 |
| miR-320e | MIMAT0015072 | 0,007 | 1,175 |
| miR-224-5p | MIMAT0000281 | 0,024 | 1,227 |
| miR-4508 | MIMAT0019045 | 0,016 | 1,369 |
| miR-575 | MIMAT0003240 | 0,014 | 1,429 |
| miR-3195 | MIMAT0015079 | 0,014 | 1,432 |
| miR-630 | MIMAT0003299 | 0,001 | 2,272 |
| Gene . | Accession # . | P value . | Fold-Change (Log2) . |
|---|---|---|---|
| miR-150-5p | MIMAT0000451 | 0,001 | -2,382 |
| let-7g-5p | MIMAT0000414 | 0,002 | -1,660 |
| miR-1260a | MIMAT0005911 | 0,009 | -1,592 |
| let-7a-5p | MIMAT0000062 | 0,010 | -1,574 |
| miR-4454 | MIMAT0018976 | 0,021 | -1,399 |
| miR-148a-3p | MIMAT0000243 | 0,030 | -1,211 |
| miR-146b-5p | MIMAT0002809 | 0,009 | -1,084 |
| miR-342-3p | MIMAT0000753 | 0,010 | -1,075 |
| let-7f-5p | MIMAT0000067 | 0,021 | -1,022 |
| miR-26a-5p | MIMAT0000082 | 0,034 | -1,008 |
| let-7d-5p | MIMAT0000065 | 0,038 | -1,005 |
| miR-30b-5p | MIMAT0000420 | 0,019 | -0,972 |
| miR-29b-3p | MIMAT0000100 | 0,044 | -0,956 |
| miR-29a-3p | MIMAT0000086 | 0,024 | -0,761 |
| miR-338-3p | MIMAT0000763 | 0,042 | 0,654 |
| miR-23a-3p | MIMAT0000078 | 0,040 | 0,689 |
| miR-222-3p | MIMAT0000279 | 0,018 | 0,756 |
| miR-548ai | MIMAT0018989 | 0,009 | 0,993 |
| miR-494 | MIMAT0002816 | 0,023 | 1,021 |
| miR-320e | MIMAT0015072 | 0,007 | 1,175 |
| miR-224-5p | MIMAT0000281 | 0,024 | 1,227 |
| miR-4508 | MIMAT0019045 | 0,016 | 1,369 |
| miR-575 | MIMAT0003240 | 0,014 | 1,429 |
| miR-3195 | MIMAT0015079 | 0,014 | 1,432 |
| miR-630 | MIMAT0003299 | 0,001 | 2,272 |
miR-150-5p Relative Expression for each subset of JMML patients with different mutational profiles and Healthy Controls
miR-150-5p Relative Expression for each subset of JMML patients with different mutational profiles and Healthy Controls
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.