Key Points
Loss of Ezh2 inhibits erythropoiesis but increases megakaryopoiesis in Jak2V617F knock-in mice.
Loss of Ezh2 induces rapid progression to myelofibrosis in mice expressing Jak2V617F.
Abstract
An activating JAK2V617F mutation has been found in ∼50% patients with myelofibrosis (MF). Inactivating mutations in histone methyltransferase enhancer of zeste homolog 2 (EZH2) also have been observed in patients with MF. Interestingly, inactivating EZH2 mutations are often associated with JAK2V617F mutation in MF, although their contributions in the pathogenesis of MF remain elusive. To determine the effects of concomitant loss of EZH2 and JAK2V617F mutation in hematopoiesis, we generated Ezh2-deficient Jak2V617F-expressing mice. Whereas expression of Jak2V617F alone induced a polycythemia vera–like disease, concomitant loss of Ezh2 significantly reduced the red blood cell and hematocrit parameters but increased the platelet counts in Jak2V617F knock-in mice. Flow cytometric analysis showed impairment of erythroid differentiation and expansion of megakaryocytic precursors in Ezh2-deficient Jak2V617F mice. Moreover, loss of Ezh2 enhanced the repopulation capacity of Jak2V617F-expressing hematopoietic stem cells. Histopathologic analysis revealed extensive fibrosis in the bone marrow (BM) and spleen of Ezh2-deleted Jak2V617F mice. Transplantation of BM from Ezh2-deleted Jak2V617F mice into wild-type animals resulted in even faster progression to MF. Gene expression profiling and chromatin immunoprecipitation sequence analysis revealed that S100a8, S100a9, Ifi27l2a, and Hmga2 were transcriptionally derepressed, and the H3K27me3 levels in these gene promoters were significantly reduced on Ezh2 deletion in hematopoietic progenitors of Jak2V617F mice. Furthermore, overexpression of S100a8, S100a9, Ifi27l2a, or Hmga2 significantly increased megakaryocytic colonies in the BM of Jak2V617F mice, indicating a role for these Ezh2 target genes in altered megakaryopoiesis involved in MF. Overall, our results suggest that loss of Ezh2 cooperates with Jak2V617F in the development of MF in Jak2V617F-expressing mice.
Introduction
Myelofibrosis (MF) is the most severe form of myeloproliferative neoplasm (MPN), characterized by deposition of fibrous tissues in the bone marrow, ineffective erythropoiesis (anemia), abnormal proliferation of megakaryocytes, and extramedullary hematopoiesis.1 Median survival of patients with MF is <6 years.2 The JAK2V617F mutation has been found in ∼50% patients with MF.3-7 Therapies targeting JAK2 kinase have been developed; however, current JAK2 inhibitors are not sufficient to produce complete remission or cure MF. This underscores the need to better understand the molecular pathogenesis of MF.
Several transgenic and knock-in mouse models of Jak2V617F have been reported.8-13 Results from these animal studies suggest that expression of Jak2V617F is sufficient to induce a polycythemia vera (PV)-like MPN. Low-grade MF is observed in some animal models of Jak2V617F after a long latency with incomplete penetrance.8-12 Therefore, additional mutations and/or genetic abnormalities might be involved in association with JAK2V617F in the pathogenesis of MF.
Enhancer of zeste homolog 2 (EZH2) is a histone methyltransferase that forms the catalytic subunit of the polycomb repressive complex 2 (PRC2). EZH2 catalyzes the methylation of histone H3 at lysine 27 (H3K27) to repress the transcription of target genes.14 EZH2 overexpression or mutations are associated with various human malignancies. EZH2 is highly expressed in prostate15 and breast16 cancers, whereas activating mutations of EZH2 (Y641 and A677 mutations) have been associated with follicular and diffuse large B-cell lymphomas.17 In contrast, inactivating mutations of EZH2 have been found in myeloid malignancies including myelodysplastic syndromes (MDS), myeloproliferative neoplasms (MPN), and MDS/MPN overlap disorders.18,19 Thus, EZH2 may have both oncogenic and tumor suppressor functions in human malignancies.
Inactivating EZH2 mutations have been observed in 6% to 13% cases of MF.1,19,20 EZH2 mutations are associated with poor survival in patients with this disease and are frequently associated with JAK2V617F,20 although their pathogenic role in MF remains unknown. To determine the contribution of inactivating EZH2 mutations in MPN mediated by JAK2V617F, we examined the effects of concomitant deletion of Ezh2 and expression of heterozygous Jak2V617F on mouse hematopoietic compartments. Our results show that deletion of Ezh2 inhibits erythropoiesis, increases megakaryopoiesis, and accelerates the development of MF in mice expressing Jak2V617F. Thus, loss of Ezh2 cooperates with Jak2V617F in the development of MF.
Materials and methods
Mice
Conditional Jak2V617F knock-in,10 Ezh2 floxed (Ezh2fl/fl),21 and MxCre22 mice were previously described. All mice were on a C57BL/6 background. Cre expression was induced by intraperitoneal injection of 5 doses of 300 μg polyinosine-polycytosine (pI-pC) at 4 weeks after birth. Wild-type C57BL/6 and BL6.SJL-Ptprca Pep3b/BoyJ (CD45.1) mice were purchased from the Jackson Laboratory. All animal studies were performed in accordance with the guidelines approved by the Institutional Animal Care and Use Committee of State University of New York (SUNY) Upstate Medical University.
Blood and tissue analysis, colony-forming assays, and flow cytometry
Blood and tissue analysis, colony-forming assays, flow cytometry, transforming growth factor-β 1 (TGF-β1) enzyme-linked immunosorbent assay, and lentiviral transduction and colony-forming unit–megakaryocyte (CFU-Mk) colony assay were performed as described in the supplemental Materials and Methods, available on the Blood Web site.
Bone marrow transplantation assays
For cell autonomous bone marrow transplantation (BMT) assay, BM (1 × 106) cells from pI-pC–induced control, Jak2VF/+, or Jak2VF/+ Ezh2−/− mice were transplanted into lethally irradiated C57BL/6 recipient mice. For competitive reconstitution assay, BM cells from uninduced control, Jak2VF/+, or Jak2VF/+ Ezh2−/− mice (CD45.2+) were mixed with CD45.1+ competitor BM cells at a 1:1 ratio and transplanted into lethally irradiated CD45.1+ congenic mice. Four weeks after transplantation, recipient animals were injected with 5 doses of pI-pC. The chimerism in the BM of transplanted animals was assessed by CD45.2 and CD45.1 expression.
Microarray analysis
Long-term hematopoietic stem cells (LT-HSCs) (Lin−Sca1+c-kit+CD34−CD135−) were sorted from the BM of control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 12 weeks after pI-pC injection using a fluorescence-activated cell sorter Aria II. Total RNA was extracted from LT-HSCs using the RNeasy Mini kit (Qiagen). RNA samples were reverse transcribed, amplified using the Ovation Pico WTA System V2 (NuGEN Technologies), and biotin labeled with the Encore Biotin Module (NuGEN Technologies). Microarray experiments were performed in the SUNY Upstate Medical University Microarray Core Facility using the Mouse Gene 2.0 ST Arrays (Affymetrix). Gene set enrichment analysis was performed as previously described.23 Microarray data and chromatin immunoprecipitation (ChIP) sequencing data have been deposited in the Gene Expression Omnibus, accession number GSE79472.
Real-time quantitative polymerase chain reaction
Total RNA was extracted from sorted LT-HSC with RNeasy Mini kit (Qiagen), and cDNA samples were prepared by using QuantiTect Reverse Transcription kit (Qiagen). Real-time quantitative PCR (RT-qPCR) was performed in a LightCycler 480 machine using SYBR Green PCR master mix (Roche Diagnostics). The data were normalized to 18S or glyceraldehyde-3-phosphate dehydrogenase, and fold changes in gene expression were determined by the ΔΔCt method. All primers used for RT-qPCR are listed in supplemental Table 1.
ChIP assay and ChIP sequencing
LSK (Lin−Sca1+c-kit+) cells were sorted from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 12 weeks after pI-pC injection. ChIP assays were performed with EpiTect ChIP OneDay Kit (Qiagen) using H3K27me3 antibody (Millipore) according to the manufacturer’s protocol. For ChIP sequencing, libraries were prepared for each sample using Illumina TruSeq ChIP Sample Prep Kit. Sequencing was carried out using the NextSeq 500 High Output Kit and NextSeq 500 next-generation sequencing instrument (Illumina). ChIP assays for human SET-2 and HEL cells were performed with EpiTect ChIP OneDay Kit (Qiagen) and H3K27me3 antibody (07-449; Millipore).
Statistical analysis
Results are presented as mean ± standard error of the mean (SEM). Statistical significance was determined by 1-way analysis of variance (ANOVA) or Student t test using GraphPad Prism, version 6 software. P < .05 was considered statistically significant.
Results
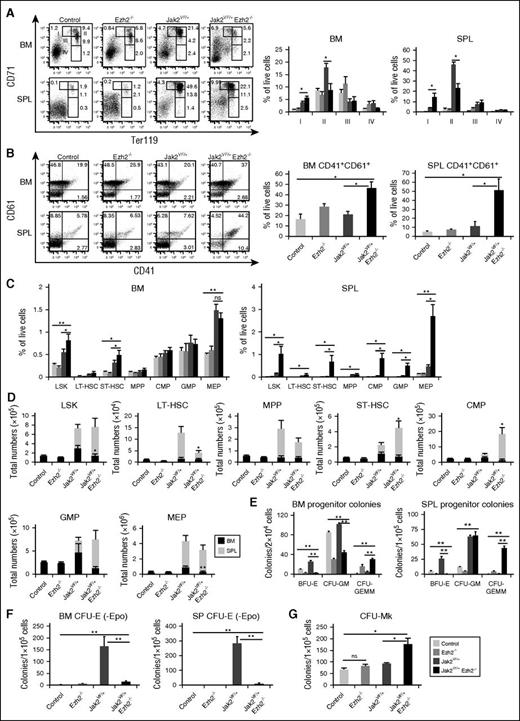
Deletion of Ezh2 inhibits erythropoiesis and promotes megakaryopoiesis in Jak2V617F knock-in mice
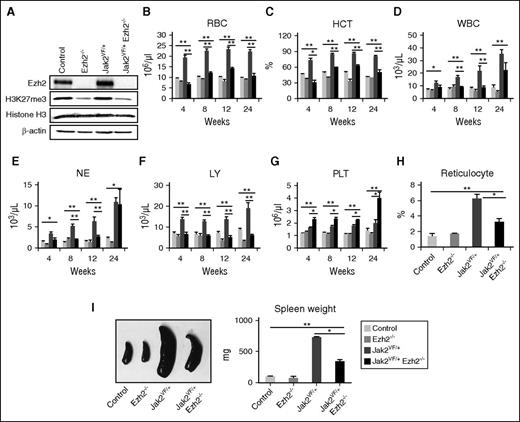
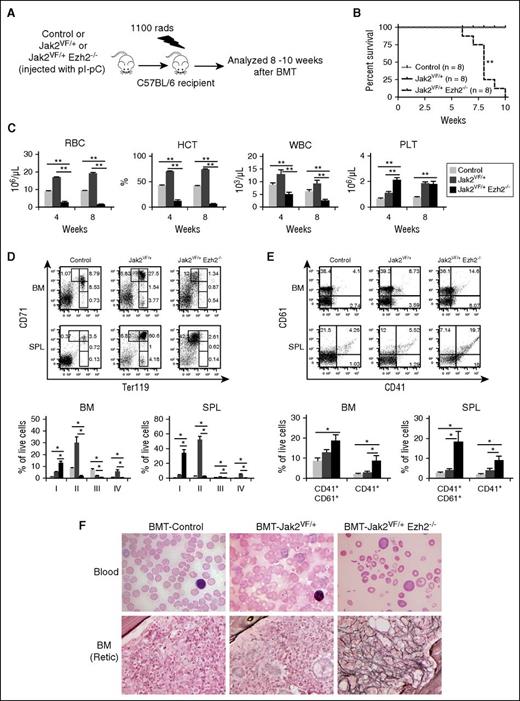
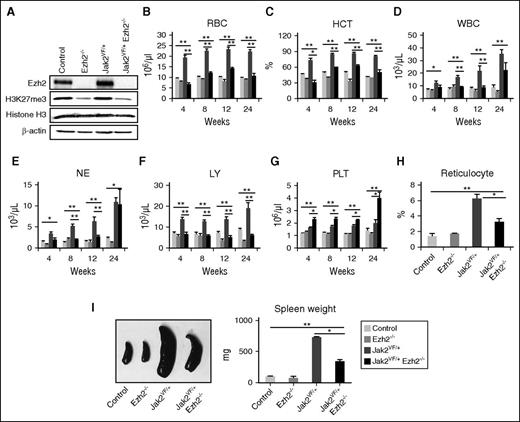
To determine the pathogenic role of concurrent inactivation of EZH2 and expression of JAK2V617F in MPN, we crossed our conditional Jak2V617F knock-in mice10 with Ezh2 floxed (Ezh2fl/fl)21 and MxCre transgenic mice,22 and generated the MxCre;Jak2V617F/+;Ezh2fl/fl mice. The expression of Jak2V617F and the deletion of Ezh2 were simultaneously induced in hematopoietic progenitors of these mice at 4 weeks after birth by pI-pC injection. Four groups of mice were analyzed: control (Ezh2fl/fl; no Cre), Ezh2 deleted (MxCre;Ezh2fl/fl referred as Ezh2−/−), Jak2V617F expressing (MxCre;Jak2V617F/+ referred as Jak2VF/+), and Ezh2 deleted Jak2V617F expressing (MxCre;Jak2V617F/+;Ezh2fl/fl referred as Jak2VF/+ Ezh2−/−) mice. As shown in Figure 1A, Ezh2 was efficiently deleted in the BM of Ezh2−/− and Jak2VF/+ Ezh2−/− mice on pI-pC induction. We also observed significant decrease in H3K27me3 levels in the BM of Ezh2−/− and Jak2VF/+ Ezh2−/− mice (Figure 1A). Consistent with our previous reports,10,24,25 Jak2VF/+ mice exhibited significantly increased red blood cells (RBCs), hematocrit, white blood cells (WBCs), neutrophils, platelets, and reticulocytes in their peripheral blood compared with control animals (Figure 1B-H). Deletion of Ezh2 significantly reduced the RBCs, hematocrit, and reticulocytes in Jak2V617F knock-in mice (Figure 1B,C,H). WBCs and lymphocyte counts were reduced, but no significant difference was observed in neutrophil counts at 24 weeks after pI-pC induction in Jak2VF/+ Ezh2−/− mice (Figure 1D-F). In contrast, platelet counts were significantly increased in Jak2VF/+ Ezh2−/− mice (Figure 1G). Mice with Ezh2 deletion alone (Ezh2−/−) exhibited modest decrease in RBCs and hematocrit but no significant alterations in platelet counts compared with control animals (Figure 1B,C,G). Splenomegaly was observed in both Jak2VF/+ and Jak2VF/+ Ezh2−/− mice (Figure 1I). The size of the spleens in Jak2VF/+ Ezh2−/− mice was smaller than in Jak2VF/+ mice but significantly larger than in control or Ezh2−/− mice (Figure 1I).
Deletion of Ezh2 inhibits erythropoiesis and promotes megakaryopoiesis in Jak2V617F knock-in mice. (A) The expression of Ezh2 and levels of H3K27me3 in the total BM cells of control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice was determined by immunoblotting at 12 weeks after pI-pC injection. β-Actin was used as a loading control. (B) RBC, (C) hematocrit (HCT), (D) WBC, (E) neutrophil (NE), (F) lymphocyte (LY), and (G) platelet (PLT) counts in the peripheral blood were assessed at 4, 8, 12, and 24 weeks after pI-pC injection in control (n = 14), Ezh2−/− (n = 9), Jak2VF/+ (n = 16), and Jak2VF/+ Ezh2−/− (n = 16) mice. (H) Reticulocytes were enumerated at 12 weeks after pI-pC injection in the peripheral blood of control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice. (I) Spleen size/weight was decreased in Jak2VF/+ Ezh2−/− mice compared with Jak2VF/+ mice, but was still significantly greater than those in control or Ezh2−/− mice (n = 5-10). One-way ANOVA was used for comparisons of all 4 groups of mice, and the Student t test was used to compare between 2 groups of mice (*P < .05; **P < .005).
Deletion of Ezh2 inhibits erythropoiesis and promotes megakaryopoiesis in Jak2V617F knock-in mice. (A) The expression of Ezh2 and levels of H3K27me3 in the total BM cells of control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice was determined by immunoblotting at 12 weeks after pI-pC injection. β-Actin was used as a loading control. (B) RBC, (C) hematocrit (HCT), (D) WBC, (E) neutrophil (NE), (F) lymphocyte (LY), and (G) platelet (PLT) counts in the peripheral blood were assessed at 4, 8, 12, and 24 weeks after pI-pC injection in control (n = 14), Ezh2−/− (n = 9), Jak2VF/+ (n = 16), and Jak2VF/+ Ezh2−/− (n = 16) mice. (H) Reticulocytes were enumerated at 12 weeks after pI-pC injection in the peripheral blood of control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice. (I) Spleen size/weight was decreased in Jak2VF/+ Ezh2−/− mice compared with Jak2VF/+ mice, but was still significantly greater than those in control or Ezh2−/− mice (n = 5-10). One-way ANOVA was used for comparisons of all 4 groups of mice, and the Student t test was used to compare between 2 groups of mice (*P < .05; **P < .005).
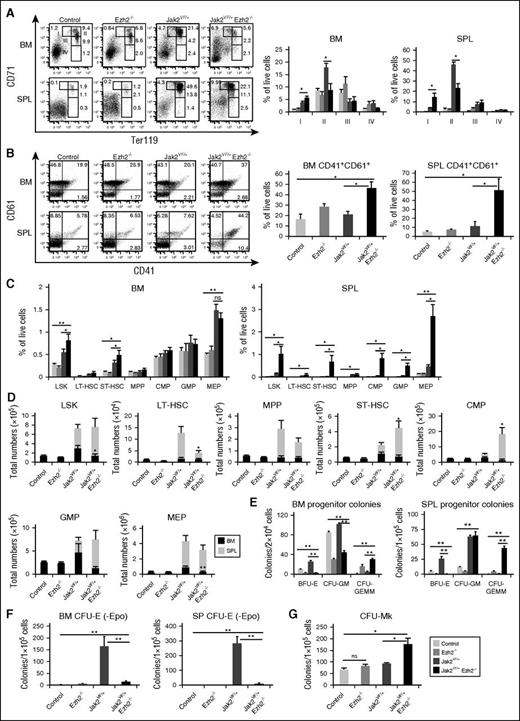
Flow cytometric analysis of the BM and spleens from Jak2VF/+ mice showed an increase in erythroid precursors (CD71+ and CD71+Ter119+) compared with control mice (Figure 2A), as we previously observed.10 Deletion of Ezh2 resulted in significant expansion of CD71high (stage I) immature early erythroid precursors but a decrease in CD71highTer119high (stage II) erythroid precursors in the BM and spleens of Jak2VF/+ Ezh2−/− mice compared with Jak2VF/+ mice (Figure 2A), suggesting that loss of Ezh2 impairs erythroid differentiation in Jak2V617F mice. The frequency of myeloid precursors (Gr-1+/Mac-1+) was comparable in the BM but was significantly increased in the spleens of Jak2VF/+ Ezh2−/− mice compared with control, Ezh2−/− and Jak2VF/+ mice (supplemental Figure 1A). The frequency of B cells (B220+) was, however, significantly reduced in the BM and spleens of Jak2VF/+ Ezh2−/− mice (supplemental Figure 1B), consistent with the previous report21 suggesting a role for Ezh2 in B-cell development. In contrast, the frequency of CD41+CD61+ megakaryocytic precursors was significantly increased in the BM and spleens of Jak2VF/+ Ezh2−/− mice compared with control, Ezh2−/−, and Jak2VF/+ mice (Figure 2B). These results suggest that concurrent loss of Ezh2 inhibits erythropoiesis but increases megakaryopoiesis in Jak2V617F mice.
Effects of Ezh2 deletion on hematopoietic precursors and progenitors in mice expressing Jak2V617F. (A) Representative dot plots of flow cytometric analysis of erythroid precursors in the BM and spleens of control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice (at 24 weeks after pI-pC injection) using surface markers CD71 and Ter119. Percentages of erythroid precursor cells at different stages of differentiation (stages I-IV, from immature to mature) are shown in bar graphs as mean ± SEM (n = 5-8). (B) Representative dot plots of flow cytometric analysis of megakaryocytic precursors in the BM and spleens of control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice using CD41 and CD61 staining. CD41+CD61+ megakaryocytic precursors in the BM and spleens are shown in bar graphs as mean ± SEM (n = 5-8). (C) Percentages of LSKs (Lin−Sca-1+c-kit+), LT-HSCs (Lin−Sca-1+c-kit+CD34−CD135−), ST-HSCs (Lin−Sca-1+c-kit+CD34+CD135−), MPPs (Lin−Sca-1+c-kit+CD34+CD135+), CMPs (Lin−Sca-1−c-kit+CD34+FcγRII/IIlow), GMPs (Lin−Sca-1−c-kit+CD34+FcγRII/IIhigh), and MEPs (Lin−Sca-1−c-kit+CD34−FcγRII/III−) in the BM and spleens from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− at 24 weeks after pI-pC are shown in bar graphs as mean ± SEM (n = 5-12). (D) Total numbers of LSKs, LT-HSCs, ST-HSCs, MPPs, CMPs, GMPs, and MEPs in the BM and spleens are shown in bar graphs as mean ± SEM (n = 5-12). (E) BM (2 × 104) or spleen cells (1 × 105) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice (n = 4-7) were plated in methylcellulose medium supplemented with cytokines. Burst-forming unit–erythroid (BFU-E), CFU-granulocyte/macrophage (CFU-GM), and CFU–granulocyte-erythroid-macrophage-megakaryocyte (CFU-GEMM) colonies were scored 7 days after plating. (F) CFU-erythroid (CFU-E) colonies. BM or spleen cells (1 × 105) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice (n = 5-7) were plated in methylcellulose medium in the absence of erythropoietin. CFU-E colonies were scored after 2 days. (G) CFU-Mk colonies. BM cells (1 × 105) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice (n = 5-7) were plated into collagen-based MegaCult medium supplemented with interleukin (IL)-3, IL-6, IL-11, and thrombopoietin. Megakaryocytic (CFU-Mk) colonies were assessed after culturing for 8 days. All data are shown as mean ± SEM. One-way ANOVA was used for comparisons of all 4 groups of mice, and the Student t test was used to compare between 2 groups of mice (*P < .05; **P < .005; ns, not significant).
Effects of Ezh2 deletion on hematopoietic precursors and progenitors in mice expressing Jak2V617F. (A) Representative dot plots of flow cytometric analysis of erythroid precursors in the BM and spleens of control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice (at 24 weeks after pI-pC injection) using surface markers CD71 and Ter119. Percentages of erythroid precursor cells at different stages of differentiation (stages I-IV, from immature to mature) are shown in bar graphs as mean ± SEM (n = 5-8). (B) Representative dot plots of flow cytometric analysis of megakaryocytic precursors in the BM and spleens of control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice using CD41 and CD61 staining. CD41+CD61+ megakaryocytic precursors in the BM and spleens are shown in bar graphs as mean ± SEM (n = 5-8). (C) Percentages of LSKs (Lin−Sca-1+c-kit+), LT-HSCs (Lin−Sca-1+c-kit+CD34−CD135−), ST-HSCs (Lin−Sca-1+c-kit+CD34+CD135−), MPPs (Lin−Sca-1+c-kit+CD34+CD135+), CMPs (Lin−Sca-1−c-kit+CD34+FcγRII/IIlow), GMPs (Lin−Sca-1−c-kit+CD34+FcγRII/IIhigh), and MEPs (Lin−Sca-1−c-kit+CD34−FcγRII/III−) in the BM and spleens from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− at 24 weeks after pI-pC are shown in bar graphs as mean ± SEM (n = 5-12). (D) Total numbers of LSKs, LT-HSCs, ST-HSCs, MPPs, CMPs, GMPs, and MEPs in the BM and spleens are shown in bar graphs as mean ± SEM (n = 5-12). (E) BM (2 × 104) or spleen cells (1 × 105) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice (n = 4-7) were plated in methylcellulose medium supplemented with cytokines. Burst-forming unit–erythroid (BFU-E), CFU-granulocyte/macrophage (CFU-GM), and CFU–granulocyte-erythroid-macrophage-megakaryocyte (CFU-GEMM) colonies were scored 7 days after plating. (F) CFU-erythroid (CFU-E) colonies. BM or spleen cells (1 × 105) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice (n = 5-7) were plated in methylcellulose medium in the absence of erythropoietin. CFU-E colonies were scored after 2 days. (G) CFU-Mk colonies. BM cells (1 × 105) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice (n = 5-7) were plated into collagen-based MegaCult medium supplemented with interleukin (IL)-3, IL-6, IL-11, and thrombopoietin. Megakaryocytic (CFU-Mk) colonies were assessed after culturing for 8 days. All data are shown as mean ± SEM. One-way ANOVA was used for comparisons of all 4 groups of mice, and the Student t test was used to compare between 2 groups of mice (*P < .05; **P < .005; ns, not significant).
Effects of Ezh2 deletion on hematopoietic stem cells/progenitors expressing Jak2V617F
We next assessed the effects of Ezh2 deletion on HSCs and progenitors in mice expressing Jak2V617F by flow cytometric analysis. The frequency of HSC-enriched LSK (Lin−Sca1+c-kit+) cells and short-term HSCs (ST-HSCs) was significantly increased in the BM and spleens of Jak2VF/+ Ezh2−/− mice compared with control, Ezh2−/−, and Jak2VF/+ mice (Figure 2C; supplemental Figure 2). Spleens of Jak2VF/+ Ezh2−/− mice also exhibited increased percentages of common myeloid progenitors (CMPs), granulocyte/macrophage progenitors (GMPs), and megakaryocyte/erythroid progenitors (MEPs) compared with Jak2VF/+ mice (Figure 2C; supplemental Figure 2). Because BM cellularity was reduced due to fibrosis in Ezh2-deleted Jak2V617F mice, the total numbers of LSK cells, GMPs, and MEPs, were decreased in the BM of Jak2VF/+ Ezh2−/− mice (Figure 2D). However, total numbers of ST-HSCs and CMPs were significantly increased in the spleens of Jak2VF/+ Ezh2−/− mice compared with control, Ezh2−/−, and Jak2VF/+ mice (Figure 2D). Also, there was a trend of increased numbers of LSK cells and GMPs in the spleens of Jak2VF/+ Ezh2−/− mice (Figure 2D).
We also preformed hematopoietic progenitor colony assays to determine the effects of Ezh2 deletion on hematopoietic progenitor compartments. The BFU-E colonies were significantly increased in the BM and spleens of Jak2VF/+ mice compared with control animals (Figure 2E). BM and spleens of Ezh2−/− and Jak2VF/+ Ezh2−/− mice showed markedly reduced BFU-E colonies but significantly increased undifferentiated mixed myeloid/erythroid CFU-GEMM colonies compared with control and Jak2VF/+ mice (Figure 2E), suggesting that the defects in erythroid differentiation in Jak2VF/+ Ezh2−/− mice could occur at an early stage of myeloid/erythroid differentiation. CFU-GM colonies in the spleens of Jak2VF/+ and Jak2VF/+ Ezh2−/− mice were comparable, although CFU-GM colonies were reduced in the BM of Jak2VF/+ Ezh2−/− mice. Erythropoietin (Epo)-independent CFU-E colonies, a hallmark feature of PV,26 was observed in the BM and spleens of Jak2VF/+ mice (Figure 2F). Deletion of Ezh2 markedly inhibited the Epo-independent CFU-E colonies in the BM and spleens of Jak2VF/+ Ezh2−/− mice (Figure 2F). We also observed marked decrease in CFU-E colonies in the presence of Epo in the BM and spleens of Jak2VF/+ Ezh2−/− mice (supplemental Figure 3). In contrast, the number of megakaryocytic (CFU-Mk) colonies was significantly increased in the BM of Jak2VF/+ Ezh2−/− mice compared with control, Ezh2−/−, and Jak2VF/+ mice (Figure 2G). The numbers of CFU-Mk colonies in the BM of Ezh2−/− mice were comparable to those in control BM (Figure 2G). Thus, concurrent loss of Ezh2 in hematopoietic progenitors of Jak2V617F mice inhibits erythroid differentiation but enhances megakaryocytic differentiation.
Loss of Ezh2 accelerates the development of myelofibrosis in Jak2V617F knock-in mice
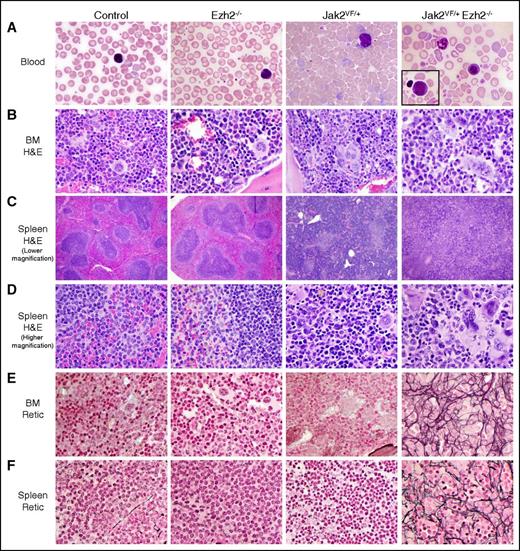
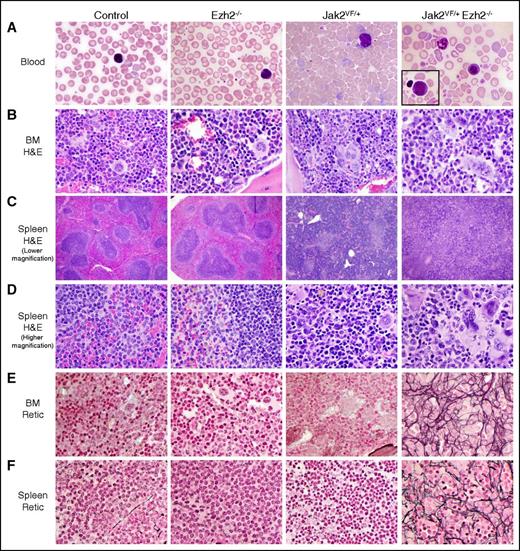
Peripheral blood smears from Ezh2−/− mice were similar to control animals (Figure 3A). Those of Jak2VF/+ mice showed marked increase in RBCs and reticulocytes (Figure 3A). Blood smears of Jak2VF/+ Ezh2−/− mice exhibited decreased RBCs and increased number of platelets (Figure 3A). BM and spleen sections from Ezh2−/− mice resembled control animals with no fibrosis (Figure 3B,E). BM of Jak2VF/+ mice showed trilineage hyperplasia with increased erythroid and myeloid precursors and increased megakaryocytes (Figure 3B). BM of Jak2VF/+ Ezh2−/− mice showed decreased erythroid precursors and increased megakaryocytes (Figure 3B). Spleen sections from Jak2VF/+ mice showed disruption of the splenic architecture with decreased white pulp and increased red pulp and with marked extramedullary trilineage hematopoiesis (Figure 3C-D). Spleens of Jak2VF/+ Ezh2−/− mice showed greater disruption of the architecture than in Jak2VF/+ mice, with prominence of megakaryocytes and myeloid precursors but fewer erythroid precursors (Figure 3C-D). Reticulin staining demonstrated extensive fibrosis (grades 2-3) in the BM and spleens of Jak2VF/+ Ezh2−/− mice at 24 weeks after pI-pC induction, whereas Jak2VF/+ mice exhibited very little or no fibrosis at this stage (Figure 3E-F). Together, these results suggest that concurrent loss of Ezh2 accelerates the development of myelofibrosis in mice expressing Jak2V617F.
Histopathologic analysis. (A) Representative peripheral blood smears (×1000 magnification) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 24 weeks after pI-pC injection are depicted. The Jak2VF/+ Ezh2−/− blood film shows leukoerythroblastic anemia, with predominance of nucleated RBC and neutrophils; a rare blast is shown in the inset. (B) Hematoxylin and eosin staining of representative BM sections (×500 magnification) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 24 weeks after pI-pC injection is shown. (C-D) Hematoxylin and eosin–stained spleen sections from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at lower (×40) and higher (×500) magnifications are presented. (E-F) Reticulin staining on representative (E) BM and (F) spleen sections (×500 magnification) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 24 weeks after pI-pC injection are shown. Note that concomitant deletion of Ezh2 in Jak2V617F mice results in extensive fibrosis in the BM and spleens, with leukoerythroblastic anemia and thrombocytosis in the blood.
Histopathologic analysis. (A) Representative peripheral blood smears (×1000 magnification) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 24 weeks after pI-pC injection are depicted. The Jak2VF/+ Ezh2−/− blood film shows leukoerythroblastic anemia, with predominance of nucleated RBC and neutrophils; a rare blast is shown in the inset. (B) Hematoxylin and eosin staining of representative BM sections (×500 magnification) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 24 weeks after pI-pC injection is shown. (C-D) Hematoxylin and eosin–stained spleen sections from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at lower (×40) and higher (×500) magnifications are presented. (E-F) Reticulin staining on representative (E) BM and (F) spleen sections (×500 magnification) from control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 24 weeks after pI-pC injection are shown. Note that concomitant deletion of Ezh2 in Jak2V617F mice results in extensive fibrosis in the BM and spleens, with leukoerythroblastic anemia and thrombocytosis in the blood.
Myelofibrosis observed in Ezh2-deficient Jak2V617F mice is transplantable
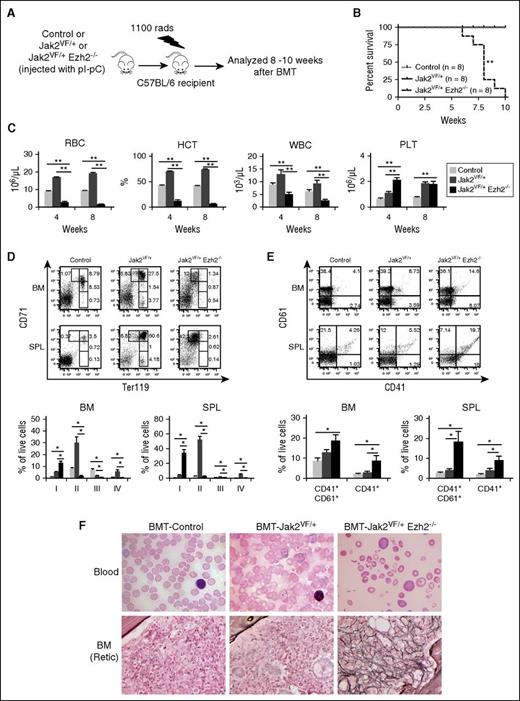
To assess whether the myelofibrosis observed in Jak2VF/+ Ezh2−/− mice was cell intrinsic, we transplanted BM cells from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice into lethally irradiated C57BL/6 wild-type recipients as outlined in Figure 4A. Recipients of Jak2VF/+ Ezh2−/− mice BM died within 7 to 10 weeks after transplantation, with a median survival of 8 weeks, whereas no death was observed in recipients of control and Jak2VF/+ BM during this time period (Figure 4B). Compared with control BM recipients, transplanted animals receiving Jak2VF/+ mice BM exhibited increased RBCs, hematocrit, WBCs, and platelets in their peripheral blood (Figure 4C). They also showed increased percentages of erythroid precursors (CD71+, CD71+Ter119+) in their BM and spleens (Figure 4D), indicating a PV-like MPN in mice transplanted with Jak2VF/+ BM cells. In contrast, recipients of Jak2VF/+ Ezh2−/− mice BM exhibited markedly reduced RBCs and hematocrit (anemia) but elevated platelet counts in their peripheral blood (Figure 4C). Recipients of Jak2VF/+ Ezh2−/− mice BM also showed increased CD71high (stage I) early erythroid precursors and decreased CD71highTer119high (stage II) erythroid precursors in their BM and spleen (Figure 4D). In contrast, the frequency of megakaryocytic precursors (CD41+ and CD41+CD61+) was significantly increased in the BM and spleens of recipients of Jak2VF/+ Ezh2−/− mice BM (Figure 4E), similar to what we observed in primary Jak2VF/+ Ezh2−/− mice (Figure 2B). The frequency of myeloid precursors (Gr-1+/Mac-1+) was comparable in the BM but was significantly increased in the spleens of transplanted animals receiving Jak2VF/+ Ezh2−/− mice BM compared with recipients of control or Jak2VF/+ mice BM (supplemental Figure 4). Peripheral blood smears of Jak2VF/+ Ezh2−/− mice BM recipients showed severe anemia (decreased RBCs, presence of teardrop RBCs), whereas recipients of Jak2VF/+ mice BM displayed polycythemia (increased RBCs and reticulocytes) (Figure 4F). Reticulin staining showed extensive bone marrow fibrosis in the recipients of Jak2VF/+ Ezh2−/− mice BM at 8 weeks after transplantation, whereas no fibrosis was detected in recipients of Jak2VF/+ mice BM at that stage (Figure 4F). Notably, recipients of Jak2VF/+ Ezh2−/− BM exhibited more severe anemia and faster progression to MF (within 8 weeks after transplantation) than the primary Jak2VF/+ Ezh2−/− mice. Together, these results suggest that the observed effects of Ezh2 loss in the development of MF in Jak2V617F mice were cell autonomous.
Effects of Ezh2 deletion on the development of MF in Jak2V617F mice are cell autonomous. (A) Experimental design for cell autonomous BMT assay. BM cells (1 × 106) were harvested from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 12 weeks after pI-pC injection and transplanted into lethally irradiated wild-type C57BL/6 recipient mice. (B) Kaplan-Meier survival analysis of transplanted animals receiving bone marrow from control (n = 8), Jak2VF/+ (n = 8), and Jak2VF/+ Ezh2−/− (n = 8) mice (**P < .005 by log-rank test). (C) Peripheral blood RBC, hematocrit (HCT), WBC, and platelet (PLT) counts were measured in recipient animals receiving control (n = 8), Jak2VF/+ (n = 8), and Jak2VF/+ Ezh2−/− (n = 9) BM at 4 and 8 weeks after BMT. (D) Flow cytometric analysis of erythroid precursors using Ter119 and CD71 surface markers in the BM and spleens of recipient mice at 8 to 10 weeks after transplantation. Representative dot plots are presented (upper). Percentages of erythroid precursors (stages I-IV, from immature to mature) are shown in bar graphs (lower) as mean ± SEM (control, n = 8; Jak2VF/+, n = 8; Jak2VF/+ Ezh2−/−, n = 7). (E) Representative dot plots (upper) and bar graphs (lower) showing percentages of megakaryocytic precursors (CD41+CD61+ and CD41+) in the BM and spleens of recipient mice. The data are presented as mean ± SEM (control, n = 8; Jak2VF/+, n = 8; Jak2VF/+ Ezh2−/−, n = 7). (F) Peripheral blood smears (×1000 magnification) from the BMT control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 8 weeks after transplantation are shown in the upper panels. Reticulin staining of representative BM sections (×500 magnification) from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− BM recipient mice at 8 weeks after BMT are shown in the bottom panels. One-way ANOVA was used for comparisons of all 3 groups of mice, and the Student t test was used to compare between 2 groups of mice (*P < .05; **P < .005).
Effects of Ezh2 deletion on the development of MF in Jak2V617F mice are cell autonomous. (A) Experimental design for cell autonomous BMT assay. BM cells (1 × 106) were harvested from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 12 weeks after pI-pC injection and transplanted into lethally irradiated wild-type C57BL/6 recipient mice. (B) Kaplan-Meier survival analysis of transplanted animals receiving bone marrow from control (n = 8), Jak2VF/+ (n = 8), and Jak2VF/+ Ezh2−/− (n = 8) mice (**P < .005 by log-rank test). (C) Peripheral blood RBC, hematocrit (HCT), WBC, and platelet (PLT) counts were measured in recipient animals receiving control (n = 8), Jak2VF/+ (n = 8), and Jak2VF/+ Ezh2−/− (n = 9) BM at 4 and 8 weeks after BMT. (D) Flow cytometric analysis of erythroid precursors using Ter119 and CD71 surface markers in the BM and spleens of recipient mice at 8 to 10 weeks after transplantation. Representative dot plots are presented (upper). Percentages of erythroid precursors (stages I-IV, from immature to mature) are shown in bar graphs (lower) as mean ± SEM (control, n = 8; Jak2VF/+, n = 8; Jak2VF/+ Ezh2−/−, n = 7). (E) Representative dot plots (upper) and bar graphs (lower) showing percentages of megakaryocytic precursors (CD41+CD61+ and CD41+) in the BM and spleens of recipient mice. The data are presented as mean ± SEM (control, n = 8; Jak2VF/+, n = 8; Jak2VF/+ Ezh2−/−, n = 7). (F) Peripheral blood smears (×1000 magnification) from the BMT control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice at 8 weeks after transplantation are shown in the upper panels. Reticulin staining of representative BM sections (×500 magnification) from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− BM recipient mice at 8 weeks after BMT are shown in the bottom panels. One-way ANOVA was used for comparisons of all 3 groups of mice, and the Student t test was used to compare between 2 groups of mice (*P < .05; **P < .005).
Deletion of Ezh2 enhances the repopulation capacity of Jak2V617F HSC and promotes expansion of myeloid lineage cells
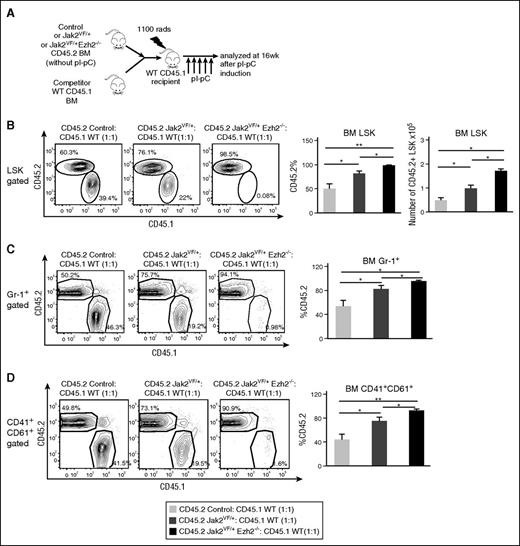
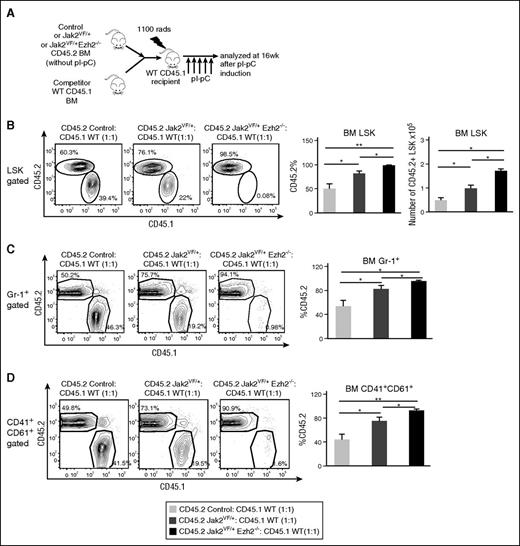
To evaluate the effects of Ezh2 loss on the function of Jak2V617F HSC in hematopoietic reconstitution and differentiation, we performed competitive repopulation assays as outlined in Figure 5A. Approximately 50% of LSK cells in the recipients of control BM were found to be donor derived (CD45.2+) (Figure 5B). Consistent with previous reports,27-29 we observed a significantly increased percentage (∼75%) of donor-derived (CD45.2+) LSK cells in recipients of Jak2VF/+ BM (Figure 5B). Recipient animals that had received Jak2VF/+ Ezh2−/− mice BM exhibited an even greater percentage (∼95%) of donor-derived (CD45.2+) LSK cells compared with the recipients of Jak2VF/+ BM (Figure 5B). The total number of donor-derived (CD45.2+) LSK cells was also significantly increased in the BM of recipients of Jak2VF/+ Ezh2−/− mice BM compared with recipients of control or Jak2VF/+ BM (Figure 5B). We also observed significantly greater percentages of donor-derived Gr-1+ myeloid and CD41+CD61+ megakaryocytic cells in recipients of Jak2VF/+ Ezh2−/− mice BM compared with control or Jak2VF/+ mice BM (Figure 5C-D). The frequency of donor-derived lymphoid cells (B220+, Thy-1+) was comparable in the BM but was significantly decreased in the spleens of recipients of Jak2VF/+ Ezh2−/− BM compared with control or Jak2VF/+ mice BM (supplemental Figure 5A-B). Together, these results suggest that loss of Ezh2 increases the repopulating capacity of Jak2V617F HSC and promotes expansion of myeloid lineage cells.
Loss of Ezh2 enhances repopulation capacity of Jak2V617F HSC and promotes myeloid cell expansion. (A) Competitive reconstitution assay. BM cells (5 × 105) from uninduced control, Jak2VF/+, or Jak2VF/+ Ezh2−/− mice (CD45.2+) were mixed with wild-type CD45.1+ mice BM (5 × 105) at a 1:1 ratio and transplanted into lethally irradiated CD45.1 recipient mice. Four weeks after BMT, recipient mice were treated with 5 doses of pI-pC to induce Ezh2 deletion and Jak2V617F expression simultaneously after hematopoietic reconstitution. The recipient mice were analyzed at 16 weeks after pI-pC injection. (B) The percentages and total numbers of donor-derived (CD45.2+) LSK cells, (C) percentages of CD45.2+ Gr-1+ myeloid cells, and (D) percentages of CD45.2+CD41+CD61+ megakaryocytic cells in the BM of recipient animals at 16 weeks after pI-pC induction are shown; bar graphs represent mean ± SEM (control, n = 5; Jak2VF/+, n = 5; Jak2VF/+ Ezh2−/−, n = 6). One-way ANOVA was used for comparisons of all 3 groups of mice, and the Student t test was used to compare between 2 groups of mice (*P < .05; **P < .005).
Loss of Ezh2 enhances repopulation capacity of Jak2V617F HSC and promotes myeloid cell expansion. (A) Competitive reconstitution assay. BM cells (5 × 105) from uninduced control, Jak2VF/+, or Jak2VF/+ Ezh2−/− mice (CD45.2+) were mixed with wild-type CD45.1+ mice BM (5 × 105) at a 1:1 ratio and transplanted into lethally irradiated CD45.1 recipient mice. Four weeks after BMT, recipient mice were treated with 5 doses of pI-pC to induce Ezh2 deletion and Jak2V617F expression simultaneously after hematopoietic reconstitution. The recipient mice were analyzed at 16 weeks after pI-pC injection. (B) The percentages and total numbers of donor-derived (CD45.2+) LSK cells, (C) percentages of CD45.2+ Gr-1+ myeloid cells, and (D) percentages of CD45.2+CD41+CD61+ megakaryocytic cells in the BM of recipient animals at 16 weeks after pI-pC induction are shown; bar graphs represent mean ± SEM (control, n = 5; Jak2VF/+, n = 5; Jak2VF/+ Ezh2−/−, n = 6). One-way ANOVA was used for comparisons of all 3 groups of mice, and the Student t test was used to compare between 2 groups of mice (*P < .05; **P < .005).
Ezh2 deletion alters gene expression in Jak2V617F HSCs
To gain insights into the mechanism by which Ezh2 deficiency promotes the development of MF in Jak2V617F knock-in mice, we performed 3 independent sets of microarray gene expression analysis on sorted LT-HSCs from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice. A heat map showed significantly upregulated and downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF LT-HSCs (Figure 6A). An analysis of microarray data revealed that 535 genes were significantly upregulated (P < .05, fold change >1.4-fold) in Jak2VF/+ Ezh2−/− LT-HSCs compared with control LT-HSCs, and 372 genes were upregulated compared with Jak2VF/+ LT-HSCs (Figure 6B). We also identified 331 genes that were significantly downregulated (P < .05, fold change <−1.4-fold) in Jak2VF/+ Ezh2−/− LT-HSCs compared with control LT-HSCs and 339 genes that were downregulated compared with Jak2VF/+ LT-HSCs (Figure 6B). A number of genes were found to be upregulated in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ LT-HSCs. These include inflammatory proteins S100a8 and S100a9, interferon pathway-related genes Ifi27l2a, Ifit1, and Cxcl10, thrombopoietin receptor Mpl, chemokines Ccl5 and Xcl1, transcription factor Pbx3, and transcriptional regulator Id2 (Figure 6C). Gene set enrichment analysis23 of microarray data revealed significant enrichment of pathways related to inflammatory responses, such as interferon α response, JNK, and tumor necrosis factor signaling pathways in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ LT-HSCs (Figure 6D). RT-qPCR further validated that expression of S100a8, S100a9, Ifi27l2a, Ifit1, Cxcl10, Mpl, Ccl5, Xcl1, Pbx3, and Id2 was significantly upregulated in Jak2VF/+ Ezh2−/− LT-HSCs compared with control or Jak2VF/+ LT-HSCs (Figure 6E).
Ezh2 deletion alters gene expression in Jak2V617F HSCs. (A) Heat map showing significantly upregulated and downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs (Lin−Sca-1+c-kit+CD34−CD135−) compared with Jak2VF LT-HSCs. (B) Venn diagrams comparing up- or downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ or control LT-HSCs. (C) A list of selected genes that are significantly upregulated in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ or control LT-HSCs. (D) Gene set enrichment analyses of the microarray data from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LT-HSCs. Enrichment plots of selected gene sets with normalized enrichment score (NES) and false discovery rate (FDR) are shown. (E) Relative expression of S100a8, S100a9, Ifi27l2a, Ifit1, Cxcl10, Mpl, Ccl5, Xcl1, Pbx3, and Id2 mRNA was determined in control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LT-HSCs by quantitative real-time PCR and normalized with 18S expression. (F) Representative histograms on cell surface expression of Mpl (thrombopoietin [TPO] receptor) in control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice BM megakaryocytes (CD41+CD61+) are shown on the left, and the fold change in median fluorescence intensity from 3 independent experiments are shown on the right as mean ± SEM. (G) Validation of some EZH2 targets in JAK2V617F-positive HEL cells. HEL cells were transduced lentiviral Ezh2 shRNA or scramble shRNA (control), and the infected cells were selected using puromycin. Relative expression of EZH2, S100A8, S100A9, IFI27 (homolog of mouse Ifi27l2a), CXCL10, CCL5, and ID2 was assessed by quantitative real-time PCR and normalized by glyceraldehyde-3-phosphate dehydrogenase. Data from 3 independent experiments are shown in bar graphs as mean ± SEM (*P < .05).
Ezh2 deletion alters gene expression in Jak2V617F HSCs. (A) Heat map showing significantly upregulated and downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs (Lin−Sca-1+c-kit+CD34−CD135−) compared with Jak2VF LT-HSCs. (B) Venn diagrams comparing up- or downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ or control LT-HSCs. (C) A list of selected genes that are significantly upregulated in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ or control LT-HSCs. (D) Gene set enrichment analyses of the microarray data from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LT-HSCs. Enrichment plots of selected gene sets with normalized enrichment score (NES) and false discovery rate (FDR) are shown. (E) Relative expression of S100a8, S100a9, Ifi27l2a, Ifit1, Cxcl10, Mpl, Ccl5, Xcl1, Pbx3, and Id2 mRNA was determined in control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LT-HSCs by quantitative real-time PCR and normalized with 18S expression. (F) Representative histograms on cell surface expression of Mpl (thrombopoietin [TPO] receptor) in control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice BM megakaryocytes (CD41+CD61+) are shown on the left, and the fold change in median fluorescence intensity from 3 independent experiments are shown on the right as mean ± SEM. (G) Validation of some EZH2 targets in JAK2V617F-positive HEL cells. HEL cells were transduced lentiviral Ezh2 shRNA or scramble shRNA (control), and the infected cells were selected using puromycin. Relative expression of EZH2, S100A8, S100A9, IFI27 (homolog of mouse Ifi27l2a), CXCL10, CCL5, and ID2 was assessed by quantitative real-time PCR and normalized by glyceraldehyde-3-phosphate dehydrogenase. Data from 3 independent experiments are shown in bar graphs as mean ± SEM (*P < .05).
Because the thrombopoietin receptor Mpl is known to play an important role in megakaryopoiesis30 and Jak2VF/+ Ezh2−/− mice exhibited enhanced megakaryopoiesis (Figure 2B), we also assessed the cell surface expression of Mpl in megakaryocytes by flow cytometry. We observed significantly increased surface expression of Mpl in megakaryocytes (CD41+CD61+) of Jak2VF/+ Ezh2−/− mice (Figure 6F).
We further validated some candidate Ezh2 target genes by knockdown of EZH2 in JAK2V617F-positive human erythroleukemia (HEL) cells using lentiviral EZH2 shRNA. We observed that knockdown of EZH2 in HEL cells resulted in significantly increased expression of S100A8, S100A9, IFI27 (homolog of mouse Ifi27l2a), CXCL10, CCL5, and ID2 (Figure 6G), suggesting that these genes are bona fide targets of EZH2.
Increased levels of TGF-β1 have been found in patients with MF, and TGF-β1 has been suggested to play a role in the pathogenesis of MF.31,32 Because transplanted animals receiving Ezh2-deleted Jak2V617F BM exhibited rapid progression to MF, we assessed the levels of TGF-β1 in these mice. We observed significantly increased levels of TGF-β1 in the serum of the transplanted animals receiving Ezh2-deleted Jak2V617F BM compared with recipients of control or Jak2V617F BM (supplemental Figure 6A). To examine if Ezh2 regulates TGF-β1 expression, we knocked down EZH2 in JAK2V617F-positive human megakaryoblastic cell line SET-2. We observed that EZH2 knockdown significantly increased the expression of TGF-β1 in SET-2 cells (supplemental Figure 6B), suggesting that loss of Ezh2 may alter the TGF-β1 expression in Jak2VF/+ Ezh2−/− mice exhibiting MF.
Ezh2 deletion alters H3K27me3 levels in LSK cells to derepress expression of Ezh2 target genes
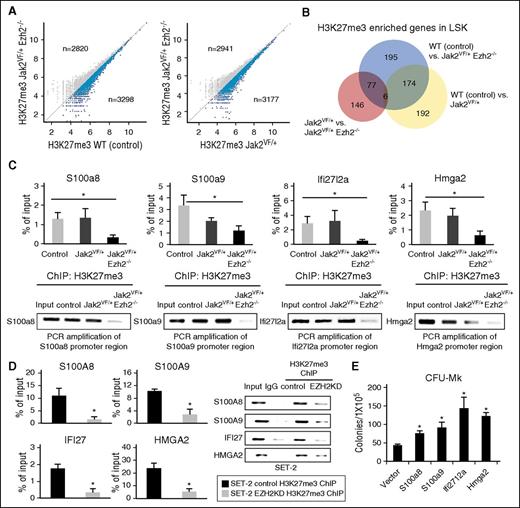
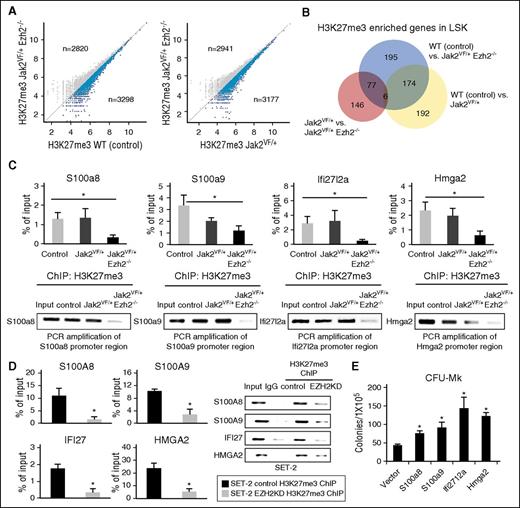
Because Ezh2 catalyzes H3K27 trimethylation (H3K27me3),14,33 deletion of Ezh2 is expected to reduce H3K27me3 levels and activate transcription of its target genes. To identify the Ezh2 target genes that are regulated by H3K27me3 in MF, we performed H3K27me3 ChIP sequencing in sorted LSK cells from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice. We observed more genes with significant (P < 1 × 10−5) H3K27me3 enrichment within 5.0 kb upstream to 0.5 kb downstream of transcriptional start sites in control or Jak2VF/+ LSK cells compared with Jak2VF/+ Ezh2−/− LSK cells (Figure 7A). We found 452 genes with significantly more H3K27me3 enrichment in control LSK cells compared with Jak2VF/+ Ezh2−/− LSK cells and 229 genes that had more H3K27me3 enrichment in Jak2VF/+ LSK cells compared with Jak2VF/+ Ezh2−/− LSK cells (P < 1.0 × 10−5; Figure 7B). We also found 372 genes had significantly less H3K27me3 enrichment in Jak2VF/+ LSK cells compared with control LSK cells (Figure 7B). ChIP followed by RT-qPCR analyses further confirmed H3K27me3 enrichment in the promoters of S100a8, S100a9, Ifi27l2a, and Hmga2 genes in control and Jak2VF/+ LSK cells and a significant reduction in H3K27me3 levels in these gene promoters in Jak2VF/+ Ezh2−/− LSK cells (Figure 7C). Consistent with this, we observed derepression of S100a8, S100a9, Ifi27l2a, and Hmga2 in multipotential progenitors (MPPs) of Jak2VF/+ Ezh2−/− mice compared with control or Jak2VF/+ MPPs (supplemental Figure 7).
Derepression of genes through alterations in H3K27me3 levels by Ezh2 deletion in Jak2V617F-positive hematopoietic cells. (A) Scatter plots showing H3K27me3 enrichment in Jak2VF/+ Ezh2−/− LSK cells compared with control or Jak2VF/+ LSK. Genes with lower H3K27me3 enrichment in Jak2VF/+ Ezh2−/− LSK cells are highlighted with blue color. (B) Venn diagram showing genes with significantly more H3K27me3 enrichment in control and Jak2VF/+ LSK compared with Jak2VF/+ Ezh2−/− LSK cells, as well as genes with significantly more H3K27me3 enrichment in control compared with Jak2VF/+ LSK cells. Genes with significant H3K27me3 enrichment in the region from 5 kb upstream to 0.5 kb downstream of the transcriptional start site are detected (P < 1.0 × 10−5). (C) H3K27me3 ChIP followed by RT-qPCR showed binding of H3K27me3 in the promoters of S100a8, S100a9, Ifi27l2a, and Hmga2 genes in control and Jak2VF/+ LSK cells. Deletion of Ezh2 significantly reduced the H3K27me3 enrichment in the promoters of these genes in Jak2VF/+ Ezh2−/− LSK cells. Results from 3 independent experiments are presented as mean ± SEM in bar graphs (*P < .05). The real-time PCR products were loaded onto 2% agarose gel. Representative pictures from agarose gel are shown in the bottom panels. (D) H3K27me3 ChIP followed by RT-qPCR showed binding of H3K27me3 in the promoters of S100A8, S100A9, IFI27, and HMGA2 genes in control and EZH2 knocked down SET-2 cells. Note that knockdown of EZH2 significantly reduced the H3K27me3 enrichment in the promoters of S100A8, S100A9, IFI27, and HMGA2 in JAK2V617F-positive SET-2 cells. The real-time PCR products were loaded onto a 2% agarose gel, and the representative pictures are shown. (E) Ectopic expression of S100a8, S100a9, Ifi27l2a, or Hmga2 significantly increased CFU-Mk colonies in the BM of Jak2VF/+ mice (n = 4-5 for each construct; *P < .05).
Derepression of genes through alterations in H3K27me3 levels by Ezh2 deletion in Jak2V617F-positive hematopoietic cells. (A) Scatter plots showing H3K27me3 enrichment in Jak2VF/+ Ezh2−/− LSK cells compared with control or Jak2VF/+ LSK. Genes with lower H3K27me3 enrichment in Jak2VF/+ Ezh2−/− LSK cells are highlighted with blue color. (B) Venn diagram showing genes with significantly more H3K27me3 enrichment in control and Jak2VF/+ LSK compared with Jak2VF/+ Ezh2−/− LSK cells, as well as genes with significantly more H3K27me3 enrichment in control compared with Jak2VF/+ LSK cells. Genes with significant H3K27me3 enrichment in the region from 5 kb upstream to 0.5 kb downstream of the transcriptional start site are detected (P < 1.0 × 10−5). (C) H3K27me3 ChIP followed by RT-qPCR showed binding of H3K27me3 in the promoters of S100a8, S100a9, Ifi27l2a, and Hmga2 genes in control and Jak2VF/+ LSK cells. Deletion of Ezh2 significantly reduced the H3K27me3 enrichment in the promoters of these genes in Jak2VF/+ Ezh2−/− LSK cells. Results from 3 independent experiments are presented as mean ± SEM in bar graphs (*P < .05). The real-time PCR products were loaded onto 2% agarose gel. Representative pictures from agarose gel are shown in the bottom panels. (D) H3K27me3 ChIP followed by RT-qPCR showed binding of H3K27me3 in the promoters of S100A8, S100A9, IFI27, and HMGA2 genes in control and EZH2 knocked down SET-2 cells. Note that knockdown of EZH2 significantly reduced the H3K27me3 enrichment in the promoters of S100A8, S100A9, IFI27, and HMGA2 in JAK2V617F-positive SET-2 cells. The real-time PCR products were loaded onto a 2% agarose gel, and the representative pictures are shown. (E) Ectopic expression of S100a8, S100a9, Ifi27l2a, or Hmga2 significantly increased CFU-Mk colonies in the BM of Jak2VF/+ mice (n = 4-5 for each construct; *P < .05).
To assess whether Ezh2 directly regulates expression of S100a8, S100a9, Ifi27l2a, and Hmga2 through H3K27me3, we performed EZH2 knockdown in JAK2V617F-positive human SET-2 and HEL cells by lentiviral EZH2 shRNA and assessed the levels of H3K27me3 in the promoters of these genes by H3K27me3 ChIP followed by RT-qPCR. We observed that knockdown of EZH2 significantly reduced H3K27me3 levels in the promoters of S100A8, S100A9, IFI27 (homolog of mouse Ifi27l2a), and HMGA2 genes in SET-2 (Figure 7D), as well as in HEL cells (supplemental Figure 8).
Because EZH2 deletion enhances megakaryopoiesis and increases CFU-Mk colonies in the BM of Jak2V617F knock-in mice, we asked if ectopic expression of Ezh2 target genes S100a8, S100a9, Ifi27l2a, and Hmga2 could increase the megakaryocytic (CFU-Mk) colony formation in the BM of Jak2V617F mice. We observed that overexpression of S100a8, S100a9, Ifi27l2a, or Hmga2 significantly increased the CFU-Mk colonies in the BM of Jak2V617F mice (Figure 7E), suggesting that derepression of these Ezh2 target genes may contribute to the enhanced megakaryopoiesis involved in MF.
Discussion
Inactivating EZH2 mutations are frequently associated with JAK2V617F mutation in MF,20 but their contributions to the pathogenesis of MF remain elusive. In this report, we demonstrate that concomitant loss of Ezh2 along with the presence of Jak2V617F promotes the development of MF. We and other groups have shown that expression of Jak2V617F in mouse hematopoietic compartments is sufficient to induce a PV-like MPN.8-12 In this study, we found that concomitant deletion of Ezh2 impairs erythropoiesis but enhances megakaryopoiesis in Jak2V617F mice. Notably, deletion of Ezh2 induces rapid progression to MF in Jak2V617F mice. Transplantation of BM from primary Ezh2-deleted Jak2V617F-expressing mice into lethally irradiated recipients resulted in even faster progression (within 8 weeks after transplantation) to MF, suggesting that the effects of Ezh2 loss and Jak2V617F expression in the development of MF are cell autonomous. Ezh2 deletion alone, however, is not sufficient to induce MF.
Several studies have suggested both oncogenic and tumor suppressor functions of Ezh2 in hematologic malignancies.17-19 Earlier studies have suggested a requirement for Ezh2 in B-cell development.21 Recent studies have shown that activating Ezh2 Y641 mutant promotes B-lymphoid transformation.34 It also has been shown that Ezh2 is required for MLL-AF9–induced leukemia.35,36 In contrast, deletion of Ezh2 in mouse hematopoietic compartments results in T-cell acute leukemia37 and MDS-like phenotype.38 Furthermore, loss of Ezh2 cooperates with Tet2 deficiency39 or RUNX1 mutation40 in the development of MDS. Interestingly, Ezh2 knock-in mice overexpressing Ezh2 in hematopoietic cells develop MPN-like disease.41 Our current study indicates that loss of Ezh2 promotes the development of MF in Jak2V617F knock-in mice. Thus, Ezh2 can positively or negatively regulate hematologic malignancies depending on the cellular context and/or associated genetic abnormalities.
We show that Ezh2 deletion enhances repopulation capacity of HSC and favors myeloid/megakaryocytic differentiation in Jak2V617F mice (Figure 5), consistent with the previous report indicating that Ezh2 deletion confers clonal advantage to the HSCs.39 In contrast, deletion of other members of PRC2 complex such as Ezh1,42 Eed,43 and Suz1244 results in decreased repopulation capacity of HSCs. These suggest that different members of the PRC2 complex have distinct roles in HSC maintenance and function.
Using gene expression profiling and RT-qPCR, we identified and validated several Ezh2 target genes (eg, S100a8, S100a9, Ifi27l2a, Ifit1, Cxcl10, Mpl, Ccl5, Xcl1, Pbx3, and Id2) that are derepressed (upregulated) in Ezh2-deficient Jak2V617F-expressing LT-HSCs (Figure 6C,E). Moreover, using ChIP-Seq and ChIP followed by RT-qPCR, we found that H3K27me3 levels were significantly reduced in the promoters of S100a8, S100a9, Ifi27l2a, and Hmga2 genes in Jak2VF/+ Ezh2−/− LSK cells (Figure 7C). We also observed derepression (upregulation) of S100a8, S100a9, Ifi27l2a, and Hmga2 genes in multipotent progenitors of Ezh2-deficient Jak2V617F (Jak2VF/+ Ezh2−/−) mice (supplemental Figure 7). Furthermore, knockdown of EZH2 significantly increased the expression of S100A8, S100A9, IFI27 (homolog of mouse Ifi27l2a), and HMGA2 and reduced the H3K27me3 levels in the promoters of these genes in JAK2V617F-positive human leukemia cells (Figures 6G and 7D). Therefore, S100a8, S100a9, Ifi27l2a, and Hmga2 are direct targets of Ezh2 that are regulated in an H3K27me3-dependent manner.
S100a8 and S100a9 are proinflammatory cytokine-like mediators that are found significantly upregulated in various human cancers including MF.45-47 A recent report has shown that increased levels of S100a8 and S100a9 block erythroid differentiation in Rps14-haploinsufficient mouse model of MDS.48 We observed erythroid differentiation defect and increased expression of S100a8 and S100a9 in Ezh2-deficient Jak2V617F (Jak2VF/+ Ezh2−/−) mice (Figures 2A and 6E). Moreover, overexpression of S100a8 or S100a9 significantly increased megakaryocytic colonies (CFU-Mk) in the BM of Jak2V617F mice (Figure 7E). Therefore, impaired erythropoiesis and enhanced megakaryopoiesis observed in Jak2VF/+ Ezh2−/− mice could be in part due to derepression (upregulation) of S100a8 and S100a9. Interferon-inducible gene IFI27 (homolog of mouse Ifi27l2a) and HMGA2 (high mobility group A2) have been found to be highly expressed in patients with MF.49,50 Moreover, ectopic expression of Hmga2 in wild-type mouse hematopoietic progenitors promotes megakaryopoiesis.51 We also observed that overexpression of Ifi27l2a or Hmga2 significantly increased megakaryocytic colonies (CFU-Mk) in the BM of Jak2V617F mice (Figure 7E). Thus, derepression of Ifi27l2a or Hmga2 evoked by the loss of Ezh2 may contribute to enhanced megakaryopoiesis involved in MF.
The canonical function of Ezh2 is to catalyze the trimethylation of H3K27.14,33 We found that Ezh2 deletion results in significant reduction in H3K27me3 levels in Jak2VF/+ Ezh2−/− LSK cells compared with control or Jak2VF/+ LSK cells (Figure 7A-B). We also observed that a significant number of genes in Jak2VF/+ Ezh2−/− LSK cells retained the H3K27me3 levels after Ezh2 deletion (Figure 7A-B). Besides Ezh2, Ezh1 can also catalyze the trimethylation of H3K27.14,52 It is thus possible that Ezh1 may partially compensate for Ezh2 loss at some of the targets of PRC2 complex.43,52
We observed increased expression of Mpl in Jak2VF/+ Ezh2−/− HSCs compared with control or Jak2VF/+ HSCs (Figure 6E). Cell surface expression of Mpl was also elevated in megakaryocytes of Jak2VF/+ Ezh2−/− mice (Figure 6F). Mpl has been shown to play an important role in megakaryopoiesis.30,53 Activating MPL mutations (MPL W515L/K) also have been associated with MF.54 It is possible that increased expression of Mpl caused by Ezh2 deletion may contribute to enhanced megakaryopoiesis in Ezh2-deficient Jak2V617F mice. However, we did not find significant change in H3K27me3 levels in Mpl gene locus among control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LSK cells. Therefore, increased expression of Mpl in Jak2VF/+ Ezh2−/− hematopoietic progenitors could be an indirect effect of Ezh2 inactivation or due to alterations in noncanonical (H3K27me3 independent) functions of Ezh2.55 Nevertheless, inactivation of Ezh2 function contributes to enhanced megakaryopoiesis and myelofibrosis in Jak2V617F knock-in mice.
In conclusion, we demonstrate that loss of Ezh2 cooperates with the Jak2V617F mutation in the pathogenesis of MF. We also provided mechanistic insights into how loss of Ezh2 promotes the development of MF in Jak2V617F mice. We show S100a8, S100a9, Ifi27l2a, and Hmga2 as direct targets of Ezh2, and derepression of these Ezh2 target genes contribute to the enhanced megakaryopoiesis involved in MF. The novel animal model of MF that we generated in this study will contribute further to our understanding of the pathogenesis of MF and facilitate in testing novel therapies for MF.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Alexandar Tarakhovsky (Rockefeller University) for providing the Ezh2 floxed mouse and Frank Middleton (SUNY Upstate Medical University) for expert help with microarray and ChIP sequencing data analysis. The authors also thank Lisa Phelps and Karen Gentile for assistance with fluorescence-activated cell sorting and processing of samples for microarray and ChIP sequencing.
This work was supported in parts by grants from the Worldwide Cancer Research (15-1381) and the National Institutes of Health, National Heart, Lung, and Blood Institute (R01 HL095685) (G.M.). G.M. is a Scholar of the Leukemia & Lymphoma Society.
Authorship
Contribution: Y.Y. performed research, analyzed data, and wrote the manuscript; H.A. and D.N. performed research; R.E.H. conducted histopathologic analysis and revised the manuscript; and G.M. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Golam Mohi, Department of Pharmacology, SUNY Upstate Medical University, 750 East Adams St, Syracuse, NY 13210; e-mail: mohim@upstate.edu.

![Figure 6. Ezh2 deletion alters gene expression in Jak2V617F HSCs. (A) Heat map showing significantly upregulated and downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs (Lin−Sca-1+c-kit+CD34−CD135−) compared with Jak2VF LT-HSCs. (B) Venn diagrams comparing up- or downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ or control LT-HSCs. (C) A list of selected genes that are significantly upregulated in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ or control LT-HSCs. (D) Gene set enrichment analyses of the microarray data from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LT-HSCs. Enrichment plots of selected gene sets with normalized enrichment score (NES) and false discovery rate (FDR) are shown. (E) Relative expression of S100a8, S100a9, Ifi27l2a, Ifit1, Cxcl10, Mpl, Ccl5, Xcl1, Pbx3, and Id2 mRNA was determined in control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LT-HSCs by quantitative real-time PCR and normalized with 18S expression. (F) Representative histograms on cell surface expression of Mpl (thrombopoietin [TPO] receptor) in control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice BM megakaryocytes (CD41+CD61+) are shown on the left, and the fold change in median fluorescence intensity from 3 independent experiments are shown on the right as mean ± SEM. (G) Validation of some EZH2 targets in JAK2V617F-positive HEL cells. HEL cells were transduced lentiviral Ezh2 shRNA or scramble shRNA (control), and the infected cells were selected using puromycin. Relative expression of EZH2, S100A8, S100A9, IFI27 (homolog of mouse Ifi27l2a), CXCL10, CCL5, and ID2 was assessed by quantitative real-time PCR and normalized by glyceraldehyde-3-phosphate dehydrogenase. Data from 3 independent experiments are shown in bar graphs as mean ± SEM (*P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/26/10.1182_blood-2015-11-679431/4/m_3410f6.jpeg?Expires=1770779232&Signature=fafBjXxcZgGYsELBxXapUcZq260oryaRjv6~XaMvu3svuN-2dzBuxuvxM7MyXjI3GxrVvvH-WxI86yp53864NuqzZxU4K77LOjlHRvbNsPGoyti4HlZFF-ctw1KUAcR-8gcSuDkEQQkSsZtbQ5~g~j2KyeQE15hrP4Oq17mVjiFqpWSkbJvD-JDP8pj8TdF189lQicHgBeyysq7XNaRK0c3aySV0O-Iq~QLamoQ4~ZV8LNDSsUe7nEQcy5rMJp-kAf8ROhZm9-EtVJlt8632rOjJmLtDRQkYjN65s6iNU6U3ji0MPD9l8VHk4hkzaQXEudG76iGhC~bmii0scHMCxg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Figure 6. Ezh2 deletion alters gene expression in Jak2V617F HSCs. (A) Heat map showing significantly upregulated and downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs (Lin−Sca-1+c-kit+CD34−CD135−) compared with Jak2VF LT-HSCs. (B) Venn diagrams comparing up- or downregulated genes in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ or control LT-HSCs. (C) A list of selected genes that are significantly upregulated in Jak2VF/+ Ezh2−/− LT-HSCs compared with Jak2VF/+ or control LT-HSCs. (D) Gene set enrichment analyses of the microarray data from control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LT-HSCs. Enrichment plots of selected gene sets with normalized enrichment score (NES) and false discovery rate (FDR) are shown. (E) Relative expression of S100a8, S100a9, Ifi27l2a, Ifit1, Cxcl10, Mpl, Ccl5, Xcl1, Pbx3, and Id2 mRNA was determined in control, Jak2VF/+, and Jak2VF/+ Ezh2−/− LT-HSCs by quantitative real-time PCR and normalized with 18S expression. (F) Representative histograms on cell surface expression of Mpl (thrombopoietin [TPO] receptor) in control, Ezh2−/−, Jak2VF/+, and Jak2VF/+ Ezh2−/− mice BM megakaryocytes (CD41+CD61+) are shown on the left, and the fold change in median fluorescence intensity from 3 independent experiments are shown on the right as mean ± SEM. (G) Validation of some EZH2 targets in JAK2V617F-positive HEL cells. HEL cells were transduced lentiviral Ezh2 shRNA or scramble shRNA (control), and the infected cells were selected using puromycin. Relative expression of EZH2, S100A8, S100A9, IFI27 (homolog of mouse Ifi27l2a), CXCL10, CCL5, and ID2 was assessed by quantitative real-time PCR and normalized by glyceraldehyde-3-phosphate dehydrogenase. Data from 3 independent experiments are shown in bar graphs as mean ± SEM (*P < .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/26/10.1182_blood-2015-11-679431/4/m_3410f6.jpeg?Expires=1770779233&Signature=KlCKC~03c71QLx1HYOrNE3YUHjGJ1kwULa5Kr7Xo8srAewL8JM7uK9cVT5ruB-KO~CPXfLdIzSx1fNf~3hAGRDvpkfl39imAqfoRiY~3iY1ehUwycfa4DdmKMLOHHKOMlnQufVjWmeW-aYeAhLX4Dnl2ixPNWSFBQ4SkzkVZG6GB2EivKdiaWjQTD~uipeiyFst4MD7arieRWyzgDdy6fZamM9O~u2ucaudKoCduUKCGyY9kN2k4GrskhYgm7u-xosNYgHLo5vWI-lkUU5ulnAGpIue4ZyI0sQVSGM0uGuEsXGMj3gxtMx7MYpLkGsRTNoQQzprDbTLT5hqmZ2zkBQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)