Key Points
HSCT recipients with multiple complement gene variants (≥3) are at high risk for severe TA-TMA.
Increased numbers of complement gene variants predisposing to TMA might contribute to racial disparities in transplant-related mortality.
Abstract
Transplant-associated thrombotic microangiopathy (TA-TMA) occurs frequently after hematopoietic stem cell transplantation (HSCT) and can lead to significant morbidity and mortality. There are no data addressing individual susceptibility to TA-TMA. We performed a hypothesis-driven analysis of 17 candidate genes known to play a role in complement activation as part of a prospective study of TMA in HSCT recipients. We examined the functional significance of gene variants by using gene expression profiling. Among 77 patients undergoing genetic testing, 34 had TMA. Sixty-five percent of patients with TMA had genetic variants in at least one gene compared with 9% of patients without TMA (P < .0001). Gene variants were increased in patients of all races with TMA, but nonwhites had more variants than whites (2.5 [range, 0-7] vs 0 [range, 0-2]; P < .0001). Variants in ≥3 genes were identified only in nonwhites with TMA and were associated with high mortality (71%). RNA sequencing analysis of pretransplantation samples showed upregulation of multiple complement pathways in patients with TMA who had gene variants, including variants predicted as possibly benign by computer algorithm, compared with those without TMA and without gene variants. Our data reveal important differences in genetic susceptibility to HSCT-associated TMA based on recipient genotype. These data will allow prospective risk assessment and intervention to prevent TMA in highly susceptible transplant recipients. Our findings may explain, at least in part, racial disparities previously reported in transplant recipients and may guide treatment strategies to improve outcomes.
Introduction
Transplant-associated thrombotic microangiopathy (TA-TMA) is a significant complication of hematopoietic stem cell transplantation (HSCT).1-5 TMA occurs when endothelial injury in the context of HSCT causes microangiopathic hemolytic anemia and platelet consumption resulting in thrombosis and fibrin deposition in the microcirculation, which affects multiple organs. By using rigorous prospective monitoring, TMA was identified in 30% to 35% of HSCT recipients and progressed to life-threatening disease in about half.3,6 The incidence of TMA is likely underestimated at many transplantation centers because some cases are mild and self-limiting and do not need active therapy, and severe TMA cases can be overlooked if appropriate laboratory tests are not performed and deaths are recorded as multisystem organ failure of unclear etiology.7
Currently, there are no data addressing individual susceptibility to TA-TMA, and there are no pretransplantation screening tools available to identify patients at risk for severe TMA. We recently reported that terminal complement activation at TMA diagnosis predicts poor survival, which suggests that complement dysregulation is a key pathway in TMA pathogenesis.8 To further explore the mechanism of TMA, we performed a hypothesis-driven prospective analysis of 17 candidate genes known to play a role in complement activation, the likely effector mechanism for vascular injury in TMA after transplantation.
Methods
Study patients
One hundred consecutive patients who underwent HSCT at Cincinnati Children’s Hospital Medical Center (CCHMC) from September 2010 to December 2011 were enrolled onto a prospective TMA biomarker study after approval from the institutional review board. Thirty-nine percent of patients met criteria for TMA using prospective monitoring as previously described.8 TMA diagnostic criteria included (1) lactate dehydrogenase above the upper limit of normal, (2) de novo thrombocytopenia with a platelet count <50 × 109/L or a ≥50% decrease in the platelet count, (3) de novo anemia with a hemoglobin below the lower limit of normal or anemia requiring transfusion support, (4) microangiopathic changes defined as the presence of schistocytes in the peripheral blood or histologic evidence of microangiopathy, and (5) absence of a coagulopathy and a negative Coombs test. ADAMTS13 activity was measured at TMA presentation to exclude diagnosis of thrombotic thrombocytopenic purpura (TTP). The date of TMA diagnosis was defined as the first date when all diagnostic criteria were fulfilled. There were 90 allogeneic transplant recipients in this study, and those with adequate genomic pretransplantation DNA (n = 77) participated in genetic analyses. Outcomes analyses to examine the impact of race on survival and occurrence of TMA were performed in an expanded cohort of the most recent 333 consecutive HSCT recipients transplanted at CCHMC. The study was conducted in accordance with the Declaration of Helsinki.
Genetic testing
Genomic DNA was isolated from recipients’ blood prior to transplantation. Seventeen genes involved in the pathogenesis of other thrombotic microangiopathies were selected for testing3,9-13 : CFH, CFHR1, CFHR3, CFHR4, CFHR5, CD55, CD59, CD46, CFI, CFB, CFP, C5, ADAMTS13, CFD, C3, C4BPA, and THBD (supplemental Table 1, available on the Blood Web site). All exons, flanking intronic and untranslated regions (5′ and 3′) were enriched or captured by using microdroplet polymerase chain reaction technology (RainDance Technologies Inc., Lexington, MA). Enriched targets were sequenced with next-generation sequencing technology on the HiSeq 2500 sequencing system (Illumina Inc., San Diego, CA) with >40-fold coverage at every target base. Resulting sequence reads were aligned against reference DNA sequence, and variants were analyzed by using NextGENe software (SoftGenetics, LLC, State College, PA). Observed variants were compared against the Single Nucleotide Polymorphism Database hosted by the National Center for Biotechnology Information. Novel variants were further evaluated by using laboratory-developed bioinformatic tools.14 Variants were filtered by focusing on coding, frame shift (indels), nonsense, and splicing modifying changes. Rare variants were defined as variants with a minor allele frequency of less than 1% in European American or African American populations based on data from the Exome Variant Server database (National Heart, Lung, and Blood Institute Exome Sequencing Project). Predicted pathogenic mutations are variants with high pathogenicity predications using in silico analyses, including SIFT, PolyPhen-2, MutationTaster, Align GVGD, and Human Splicing Finder splice site prediction algorithms. All identified variants were confirmed by Sanger sequencing (Applied Biosystems). In addition, deletions at the CFHR3/CFHR1 loci were detected by using the multiple ligation-dependent probe amplification method (MRC Holland, Amsterdam, The Netherlands). For completeness, we also included synonymous variants. These sequence variants that were otherwise neutral for amino acids could create a cryptic splicing site and represent unique haplotypes that are important in susceptibility to TMA or they could regulate transcription or translation.
RNA-Seq analysis
RNA sequencing (RNA-Seq) was performed by Genomics, Epigenomics and Sequencing Core at the University of Cincinnati on pretransplantation whole-blood mononuclear cells from 8 HSCT recipients with TMA who had identified gene variants and 8 HSCT recipients without TMA and without any identified gene variants based on sample availability. We selected pretransplantation RNA (presumed resting state) for this assay because we wanted to examine the impact of genotype, not the possible effects of activation of complement during the transplantation process. Expression of each gene with a genetic variant and of pathways related to complement activation were compared with expression of the same gene in HSCT patients with and without TMA.
The RNA sequencing of 50 nucleotide long single-end reads was performed by using the standard Illumina protocol on the Illumina HiSeq system. Sequence reads were aligned to the reference human genome by using the TopHat aligner,15 and reads aligning to each known transcript were counted by using Bioconductor packages for next-generation sequencing data analysis.16 The differential expression analysis between TMA samples with variants in a specific gene and non-TMA samples without variants was performed for each gene separately based on the negative-binomial statistical model of read counts as implemented in the Bioconductor DESeq package.17 The pathway enrichment analysis was performed separately for upregulated and downregulated genes by using the LRpath methodology18 and the MSigDB database of gene lists and pathways.19 The enrichment P values were adjusted for testing all pathways in MSigDB using the false discovery rates.20
Statistical analysis
Median (range) and frequencies were reported to describe continuous and categorical variables, respectively. Differences by group for continuous and categorical variables were compared by using Fisher exact and Wilcoxon tests, respectively. Survival and competing risk analyses were performed by using the Kaplan-Meier method. Log-rank tests were used to assess the difference in overall survival by group. Gray’s method was used to test for difference in cumulative incidence by group. For cumulative incidences, death by relapse or all-cause mortality were considered competing risks in the analyses of nonrelapse mortality and TMA, respectively. Analyses were performed by using R version 3.1.3. All statistical tests were two-sided, and significance was assessed at P < .05.
Results
Study demographics and patient characteristics
Seventy-seven of the 90 allogeneic transplant recipients enrolled onto the study had adequate pretransplantation DNA for analysis. Thirty-four of these HSCT recipients had a clinical diagnosis of TMA based on prospective screening, and 43 did not have TMA. Study cohort characteristics are summarized in Table 1. Study participants were children younger than 18 years of age, and the majority were white males. A majority of patients (66%) had HSCT for nonmalignant disorders, including bone marrow failure or immunodeficiency. The stem cell source was mainly bone marrow (78%) from unrelated donors (73%). A similar number of patients received myeloablative (53%) and reduced-intensity (47%) regimens. Even though nonwhite transplant recipients composed only 18% of our total study cohort, a significantly higher fraction of nonwhites than whites had TMA (71% vs 39%; P = .04). All transplant recipients received cyclosporine (CSA) for graft-versus-host disease (GVHD) prophylaxis. Uniform drug monitoring was used to maintain a CSA trough of 250 to 350 ng/mL. There was no difference in CSA levels between patients who developed TMA and those who did not (251.6 and 247.2 ng/mL, respectively; P = .65). Children with Fanconi anemia had increased incidence of TMA. These were the only patients who received both T-cell–depleted peripheral blood stem cells and CSA, so we were not able to tell whether the increased risk of TMA reflected increased susceptibility in persons with Fanconi anemia or was associated with the treatment protocol itself. Similarly, children with diagnoses of malignant disease received CSA and methotrexate for GVHD prophylaxis. Both malignancy and receiving methotrexate were associated with lower incidence of TMA, so we could not determine whether diagnosis or treatment regimen influenced the incidence of TMA. None of the study patients received any targeted therapy (eg, complement blockade) for TMA.
Gene variants identified
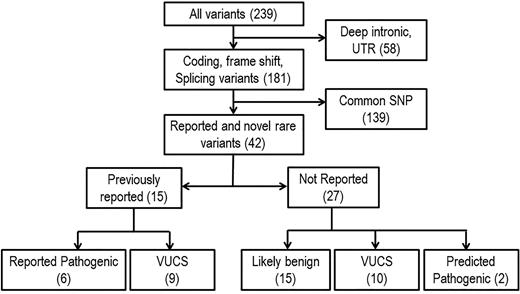
A total of 239 variants in 17 genes were identified after initial quality control (>40-fold coverage and elimination of false positives by visual inspection and Sanger sequencing). Three genes (CFP, C4BPA, and CD59) were not further considered because no variants were identified. After excluding deep intronic and untranslated region variants (n = 58) and common single nucleotide polymorphisms (n = 139), we identified 42 likely functional variants, including 15 previously reported microangiopathy-related variants. Of the 15 previously reported variants, 6 are known to be pathogenic, and 9 are variants with uncertain clinical significance. Of the 27 previously unreported variants, 2 are predicted to be pathogenic, 10 are considered variants with uncertain clinical significance, and 15 are predicted as possibly benign by computer methods for predicting functional consequences of sequence variants (Figure 1). We performed pathogenicity predictions using SIFT, PolyPhen-2, MutationTaster, Align GVGD, and Human Splicing Finder splice site prediction algorithms to evaluate the 27 variants not previously described (2 nonsense, 9 missense, 12 synonymous, and 3 splice site variants). Two nonsense variants in CFHR1 and CFHR3 were considered pathogenic by the nature of predicted effects, and 15 variants, including synonymous and splice site variants, were predicted as possibly benign (supplemental Table 2). Because predictive algorithms are of limited accuracy, considered by many to be correct only about 70% of the time, we did not want to overlook important changes for variants predicted as possibly benign, so these were retained in the analysis, and gene expression profiling was performed on available samples (see “RNA-Seq analysis”).
Gene filtering schema. Variants were filtered by focusing on coding and noncoding functional modifying changes. Rare variants are defined as variants with a minor allele frequency less than 1% in European American or African American populations based on data from the Exome Variant Server database (National Heart, Lung, and Blood Institute Exome Sequencing Project). Predicted pathogenic mutations are rare nonsense mutations, indels, or missense variants with highly positive pathogenicity predications using SIFT, PolyPhen-2, MutationTaster, and Align GVGD. SNP, single nucleotide polymorphism; UTR, untranslated region; VUCS, variant with uncertain clinical significance.
Gene filtering schema. Variants were filtered by focusing on coding and noncoding functional modifying changes. Rare variants are defined as variants with a minor allele frequency less than 1% in European American or African American populations based on data from the Exome Variant Server database (National Heart, Lung, and Blood Institute Exome Sequencing Project). Predicted pathogenic mutations are rare nonsense mutations, indels, or missense variants with highly positive pathogenicity predications using SIFT, PolyPhen-2, MutationTaster, and Align GVGD. SNP, single nucleotide polymorphism; UTR, untranslated region; VUCS, variant with uncertain clinical significance.
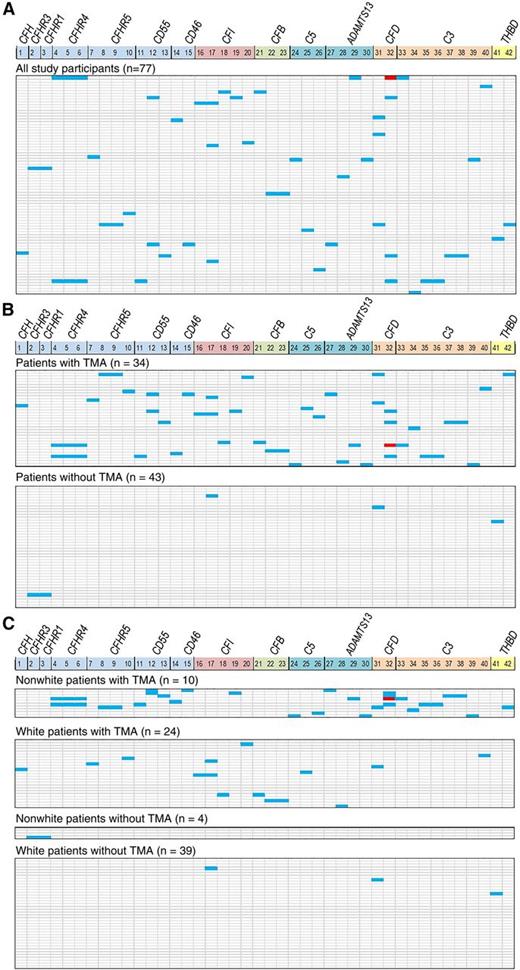
Thirty-four percent of tested patients (26 of 77) had at least one gene variant identified (Figure 2A). Figure 2B displays a heat map of gene variant distribution stratified according to the presence or absence of TMA after HSCT and shows significantly higher frequency of variants in patients with TMA. Twenty-two (65%) of 34 patients with TMA had at least 1 gene variant identified compared with 4 (9%) of 43 patients without TMA (P < .0001). Pathogenic variants previously described in other microangiopathies were seen in CFHR5, CFI, and ADAMTS13, all in transplant recipients with TMA. ADAMTS13 activity tested at TMA diagnosis was slightly reduced (43% to 50%) in patients with ADAMTS13 variants but did not meet the criteria for diagnosis of TTP. Three CFI variants seen in our study have previously been reported in acute hemolytic-uremic syndrome (aHUS) and 2 ADAMTS13 variants were reported in TTP; 1 CFHR5 variant is known to cause CFH-related protein deficiency.21-25 A CFD variant (c.357+16C>A) was detected only in African American patients with TMA. The median number of gene variants seen in transplant recipients with TMA was 1 (range, 0-7) and 0 (range, 0-2) in those without TMA (P < .0001). In addition to results from next-generation sequencing, 2 recipients with TMA had homozygous deletion of CFHR3/CFHR1 detected by multiple ligation-dependent probe amplification that was not seen in recipients without TMA.26 Both of the patients with homozygous deletion of CFHR3/CFHR1 had severe multivisceral TMA and died. No known pathogenic variants were seen in recipients without TMA. The high frequency of variants in recipients with TMA compared with those without TMA (distributed by the selective pressure of transplantation) suggests that the variants are associated with TMA and do not represent random polymorphic variation in the genes tested. Analysis with all variants predicted by computer algorithm as possibly benign excluded did not change the conclusions of the study (supplemental Figure 1).
Gene variants. Gene variants identified after a rigorous filtering process are shown as a heat map. Each vertical column marks 14 genes that were included after data filtering, some with multiple variants (CFP, C4BPA, and CD59 were excluded from the 17-gene panel because of the absence of the variants detected). Numerical list 1-42 correlates with the specific gene variant listed in supplemental Table 2. Each horizontal line represents 1 study patient (HSCT recipient). Heterozygous variants are shown in blue and homozygous variants are shown in red. (A) Gene variants identified in all 77 HSCT recipients: 26 (34%) of 77 of patients had at least 1 gene variant detected. (B) A majority of gene variants are present in HSCT recipients with TMA (top panel), whereas few variants are seen in those without TMA (bottom panel). The median number of gene variants (of known or uncertain clinical significance) seen in recipients with TMA was 1 (range, 0-7) and 0 (range, 0-2) in those without TMA (P < .0001). (C) Gene variant distribution by race and TMA. All 10 nonwhites (100%) with TMA had at least 1 variant detected, whereas 12 (50%) of 24 of whites had variants identified. Three or more (range, 3-7) variants were identified only in nonwhite recipients with TMA (n = 7); 5 (71%) of these 7 patients died of severe TMA, indicating the effect of race and complement gene variants on TMA phenotype. CFD variant c.357+16C>A (number 32) was identified exclusively in African American HSCT recipients. Of 12 whites with TMA who had gene variants identified, only 3 had 2 variants and the other 9 had a single variant detected. Only 3 (7.7%) of 39 nonwhites without TMA had a single variant detected.
Gene variants. Gene variants identified after a rigorous filtering process are shown as a heat map. Each vertical column marks 14 genes that were included after data filtering, some with multiple variants (CFP, C4BPA, and CD59 were excluded from the 17-gene panel because of the absence of the variants detected). Numerical list 1-42 correlates with the specific gene variant listed in supplemental Table 2. Each horizontal line represents 1 study patient (HSCT recipient). Heterozygous variants are shown in blue and homozygous variants are shown in red. (A) Gene variants identified in all 77 HSCT recipients: 26 (34%) of 77 of patients had at least 1 gene variant detected. (B) A majority of gene variants are present in HSCT recipients with TMA (top panel), whereas few variants are seen in those without TMA (bottom panel). The median number of gene variants (of known or uncertain clinical significance) seen in recipients with TMA was 1 (range, 0-7) and 0 (range, 0-2) in those without TMA (P < .0001). (C) Gene variant distribution by race and TMA. All 10 nonwhites (100%) with TMA had at least 1 variant detected, whereas 12 (50%) of 24 of whites had variants identified. Three or more (range, 3-7) variants were identified only in nonwhite recipients with TMA (n = 7); 5 (71%) of these 7 patients died of severe TMA, indicating the effect of race and complement gene variants on TMA phenotype. CFD variant c.357+16C>A (number 32) was identified exclusively in African American HSCT recipients. Of 12 whites with TMA who had gene variants identified, only 3 had 2 variants and the other 9 had a single variant detected. Only 3 (7.7%) of 39 nonwhites without TMA had a single variant detected.
In patients of our study cohort evaluated for genetic variants (n = 77), the cumulative incidence of TMA was higher in nonwhites than in whites (71% vs 39%; P = .04). Moreover, more gene variants were detected in nonwhite than in white transplant recipients: 2.5 (range, 0-7) vs 0 (range, 0-2) (P < .0001). Figure 2C provides a heat map of gene variants stratified by race and TMA, illustrating that the highest number of variants occurred in nonwhite patients with TMA.
Seven patients with TMA, all nonwhites, each had between 3 and 7 variants identified (3 patients had 3 variants each, 2 had 4 variants each, and 2 had 7 variants each). Five of these 7 nonwhite patients with ≥3 gene variants died of severe TMA, indicating an important association of race and cumulative effect of complement gene variants with TMA phenotype. Only 1 of the 4 nonwhite recipients without TMA had 2 variants detected. None of the white recipients without TMA had more than 2 variants detected.
To address whether gene variants were associated with race vs TMA, we analyzed only white HSCT recipients. Fifty percent of whites (12 of 24) with TMA had at least 1 variant compared with 7.7% of whites (3 of 39) without TMA. Eighty percent of white patients with at least 1 gene variant (12 of 15) developed TMA compared with 25% of white patients (12 of 48) without any variant (P < .0001). This observation indicates that the presence of gene variants is significantly enriched in patients of any race who develop TMA, but nonwhites have more variants and more severe disease.
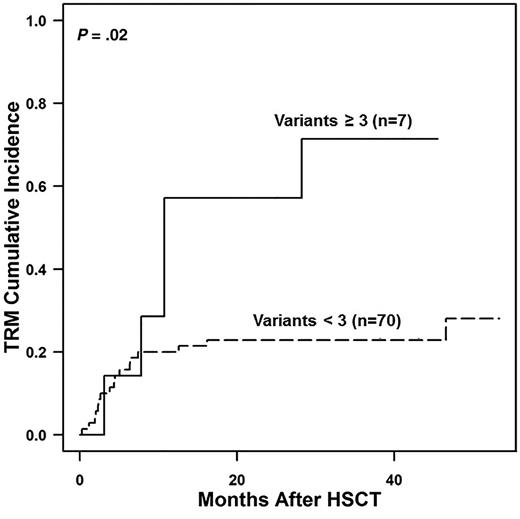
Next, we compared outcomes in recipients who had 0 to 2 or ≥3 gene variants. Patients with ≥3 gene variants, all of whom were nonwhites, had increased transplant-related mortality (TRM) (57% vs 21% at 1 year; P = .02) (Figure 3). These data led us to hypothesize that TRM would be elevated and survival would be reduced in nonwhites who received transplantations at our center. We analyzed an expanded cohort of 333 consecutive transplantations performed at CCHMC comparing survival in patients with and without TMA in white and nonwhite recipients. This larger cohort included 48 nonwhite recipients, most of whom (85%) were African Americans. The cumulative incidence of TMA in this cohort was 35% (40% in allogeneic transplant recipients and 8% in autologous transplant recipients; P < .0001) and again was significantly more frequent in nonwhite than in white HSCT recipients (52% vs 29%; P = .008). Supporting our hypothesis, nonwhite recipients had worse survival than white recipients (56% vs 81% at 1 year; P = .0009), and HSCT recipients with TMA had inferior survival compared with those without TMA (63% vs 85% at 1 year; P = .0001). Furthermore, nonwhite patients with TMA had significantly worse survival after transplantation than white patients with TMA (37% vs 69% at 1 year; P = .04), whereas survival was not different by race in patients without TMA (73% vs 87% at 1 year; P = .12), supporting our hypothesis that at least a proportion of the inferior survival seen in nonwhite HSCT recipients is mediated by increased susceptibility to severe TMA (Figure 4).
Transplant-related mortality (TRM) in relation to gene variant number identified. Shown is TRM in patients with 0 to 2 gene variants detected compared with those with ≥3 gene variants detected. TRM among study patients was calculated by using the Kaplan-Meier method. The 2 groups were found to significantly differ by using the log-rank test (P = .02). The TRM at 1 year and 3 years after HSCT was much higher in patients with ≥3 gene variants (57% at 1 year and 71% at 3 years) compared with the patients with 0 to 2 variants (21% at 1 year and 24% at 3 years).
Transplant-related mortality (TRM) in relation to gene variant number identified. Shown is TRM in patients with 0 to 2 gene variants detected compared with those with ≥3 gene variants detected. TRM among study patients was calculated by using the Kaplan-Meier method. The 2 groups were found to significantly differ by using the log-rank test (P = .02). The TRM at 1 year and 3 years after HSCT was much higher in patients with ≥3 gene variants (57% at 1 year and 71% at 3 years) compared with the patients with 0 to 2 variants (21% at 1 year and 24% at 3 years).
TMA and race effect on survival among patients who received a transplant. Kaplan-Meier survival curves (total n = 333). HSCT survival is inferior in (A) HSCT patients who developed TMA (63% vs 85% 1-year overall survival [OS] and 52% vs 83% 3-year OS; P = .0001) and in (B) nonwhite HSCT recipients (56% vs 81% 1-year OS and 56% vs 75% 3-year OS; P = .0009). (C-D) Effect of TMA and race combined on HSCT survival. (C) Race did not have significant influence on survival in HSCT recipients without TMA (73% vs 87% 1-year OS and 73% vs 84% 3-year OS; P = .12). (D) Nonwhite HSCT recipients who developed TMA had significantly inferior survival compared with whites who developed TMA (37% vs 69% 1-year OS and 37% vs 55% 3-year OS, P = .04).
TMA and race effect on survival among patients who received a transplant. Kaplan-Meier survival curves (total n = 333). HSCT survival is inferior in (A) HSCT patients who developed TMA (63% vs 85% 1-year overall survival [OS] and 52% vs 83% 3-year OS; P = .0001) and in (B) nonwhite HSCT recipients (56% vs 81% 1-year OS and 56% vs 75% 3-year OS; P = .0009). (C-D) Effect of TMA and race combined on HSCT survival. (C) Race did not have significant influence on survival in HSCT recipients without TMA (73% vs 87% 1-year OS and 73% vs 84% 3-year OS; P = .12). (D) Nonwhite HSCT recipients who developed TMA had significantly inferior survival compared with whites who developed TMA (37% vs 69% 1-year OS and 37% vs 55% 3-year OS, P = .04).
We evaluated the functional importance of genetic variants using RNA-Seq and compared gene expression in peripheral blood mononuclear cells collected before transplantation from 8 transplant recipients with high numbers of gene variants and TMA and 8 recipients who did not have any variants identified and did not develop TMA. The pathway analysis of differentially expressed genes (Table 2) indicated upregulation of complement-associated pathways in patients with variants compared with those without variants. Importantly, we observed upregulation of complement pathways even in patients with variants predicted as possibly benign by some algorithms. Of particular interest, a C5 variant (c.921G>C [#26]) that was predicted to be benign was the only detected variant in the patient with TMA, and it showed upregulation of multiple complement pathways, which supports a possible role in disease. The analysis provided no evidence of downregulation of complement pathways in any patients with gene variants. These data support our hypothesis that identified gene variants may facilitate rapid complement activation.
Discussion
We report a hypothesis-driven analysis of 17 candidate genes that participate in complement activation, a possible effector mechanism for vascular injury in HSCT-associated TMA. This is the first large prospective study of genetic susceptibility to TMA after HSCT. We identified gene variants in 65% of HSCT recipients with TMA compared with only 9% in those without TMA. TMA incidence was higher in patients with gene variants detected regardless of race. However, TMA incidence and number of gene variants were higher in nonwhite transplant recipients, the majority of whom were African Americans, compared with white recipients. Importantly, the higher number of variants observed in nonwhite HSCT recipients was associated with a more severe TMA and high mortality.
Our data support the hypothesis that complement gene variants modify susceptibility to TMA after HSCT. We believe that the occurrence of TMA requires at least 2 factors for sufficient vascular injury (often caused by drugs such as chemotherapy or calcineurin inhibitors) in a genetically susceptible host. It is likely that additional and as yet poorly defined environmental stressors such as viral infections and GVHD contribute to this gene-environment interaction and to which children develop TMA. In any one person, the relative contribution of genetic susceptibility and environmental stressors may be different, so strong environmental pressure may cause TMA in a relatively genetically resistant person, and conversely, little environmental stress might be needed to cause TMA in a highly susceptible person. We therefore propose that genotype can serve clinically as an indicator of persons at increased risk for TMA after transplantation because additional environmental stressors are needed for the phenotype of TMA. We believe that the identification of persons at increased risk of TMA has clinical utility. Most simply, genotype might serve to identify persons in whom surveillance for complement activation and early intervention with complement blockers might be indicated. Those who perform transplantations might consider use of a transplantation strategy for persons with multiple variants that reduces environmental stressors such as transplantation using an ex vivo T-cell–depleted product that removes the need for calcineurin inhibitors and prevents the occurrence of GVHD.
We do not believe that the high frequency of variants in nonwhites simply represents inconsequential increased genetic polymorphism in these populations, because we observed a clear association of increased variants with increased incidence and severity of TMA in any race, although nonwhites had more variants and more severe disease phenotype. Moreover, RNA-Seq studies support increased complement activation in transplant recipients with genetic variants but not in transplant recipients without TMA and and with no identified genetic abnormalities, which indicates likely functional importance for the variants. Additional functional assays that examine complement activation and deposition on vascular endothelium in persons with and without complement gene variants are under way and will likely aid in identification of variants associated with a severe TMA phenotype.
We propose that our data indicate that HSCT-associated TMA is driven by complement-mediated tissue injury and that complement dysregulation is not simply a marker of tissue injury incurred during transplantation. We recognize that separating these 2 possibilities can be challenging. Perhaps the best indicator that complement activity is causative of TMA would be the demonstration that blocking complement activity improves outcomes. Although none of the patients in this particular cohort received any targeted therapy for TMA, in subsequent work, we treated 18 patients with high-risk TMA and evidence of terminal complement activation with the complement blocker eculizumab. Sixty-one percent of patients achieved complete resolution of TMA with eculizumab therapy. Overall survival in this group was 56% at 1 year after TMA diagnosis, whereas these patients historically had very poor survival of 9% (P = .003), as we show in our recent prospective study.27 The good response to complement blockade that we observed in patients with severe TMA supports our current hypothesis that complement system dysregulation plays an important role in TMA pathogenesis, and the complement system is a potential therapeutic target for this disease. Future studies will be needed to examine the response to complement blocking therapy in patients with different complement gene variants.
Our transplant population includes more children with nonmalignant disorders than is typical of most transplantation centers, reflecting the clinical and research focus of our program. There were too few patients with each diagnosis to determine whether the high frequency of variants was being driven by children with particular diagnoses. Future studies are needed to determine whether our data are generalizable to pediatric and adult transplantation programs with greater focus on malignancies.
Our data indicate that HSCT-associated TMA falls under the umbrella of complement-mediated disorders, but it has unique genetic and clinical features separating it from other complement diseases such as aHUS. The number of persons with TA-TMA likely exceeds the total number with other rare forms of microangiopathies. The predictable occurrence of TA-TMA in large cohorts of patients, as opposed to sporadic and unpredictable occurrence in other microangiopathies, offers a unique opportunity to study the evolution of a microangiopathy prospectively. Previous studies have identified genetic variants in complement genes in about 60% of persons with aHUS.9 There are no reports that identify the large number of variants in individual persons that we found in our analysis (up to 7).28 This difference may be the result of lack of comprehensive genotyping in prior studies, or the small number of persons of African descent in the primarily European-based reports.
The finding of multiple variants occurring at high frequency in persons of African descent in our cohort leads us to hypothesize that there is a selective benefit of rapid complement activation in Africans.29 Fast complement activation is needed in response to infections like Neisseria meningitidis, which raises the possibility that variants observed exclusively in patients of African descent might allow rapid complement activation and provide protection against meningitis, a prevalent cause of mortality in Africa.30-32 In support of this, recent reports document complement variants that are protective against Neisseria meningitidis in African patients.29 In areas where meningitis is endemic (African meningitis belt), fast complement activation would serve as a protective feature. But such genetic makeup may be harmful during stem cell transplantation and could result in significant tissue damage from uncontrolled complement activation with multiple triggers such as chemotherapy, infections, and GVHD that occurs after HSCT and results in severe vascular injury and TMA. It has also been documented in other clinical settings that persons who require a low trigger for complement activation are less prone to infections but are more likely to suffer from inflammatory conditions.29 Our RNA-Seq data confirm upregulation of multiple complement pathways in identified variants, including those predicted as possibly benign in individuals of African descent, and this likely allows rapid complement activation under stress during transplantation. We recognize that each of the many variants described in this study may have greater or lesser impact on complement activity, and perhaps the cumulative genotype may drive phenotype rather than any single variant. We also recognize that some of the variants may not have functional importance, and we are hopeful that our ongoing functional complement deposition studies will help decipher this.
Our data show increased TRM and reduced survival in nonwhite transplant recipients, especially those with TMA. A previous registry report described inferior survival in African Americans after unrelated donor HSCT, but it was unable to attribute reduced outcomes to risk factors such as HLA-matching and socioeconomic status.33 Our study identified the importance of individual genetic susceptibility to TMA and the significant impact of TMA on survival, perhaps explaining, at least in part, racial disparities in HSCT outcomes.
Our work brings important and novel insights into the potential genetic predisposition to TMA after HSCT that may have an immediate therapeutic importance. We believe that some of our identified genetic variants may not have biological importance in the course of normal life, but under the stress of HSCT, with significant vascular injury caused by intense radiation and/or chemotherapy, susceptible individuals will likely manifest TMA. Pretransplantation genetic screening alone or in combination with functional tests will allow selection of high-risk HSCT recipients who could benefit from specific interventions, because HSCT is a planned therapeutic procedure and transplantation strategies can be modified accordingly. Calcineurin inhibitors and sirolimus are associated with increased risk of TMA. Alternative strategies for control of acute GVHD (eg, ex vivo T-cell depletion or use of photopheresis as prophylaxis) could be used, and novel complement blockers might be a therapeutic option in high-risk patients. Close monitoring of high-risk patients and early therapeutic interventions before organ damage occurs should improve overall transplantation outcomes. With improved understanding of the pharmacokinetics and pharmacodynamics of eculizumab in this clinical setting and the use of TMA biomarkers that predict poor outcome, therapy can be initiated earlier with more reliable complement blockade that likely will result in further improvement of therapy response. These data also help us understand mechanisms of disease that will be relevant to other microangiopathies for which populations are smaller and prospective systematic study is not possible.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge Dr Jörg Köhl for valuable assistance with interpretation of complement analyses.
This work was supported in part by an unrestricted research grant from Alexion Pharmaceuticals. Alexion had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Authorship
Contribution: S.J., B.L., and S.M.D. designed the study, performed research, and wrote the paper; K.Z. and F.Z. performed genetic testing and analysis; J.M., M.M., and J.C. performed functional studies and analysis; A.L. performed statistical analyses; K.C.M. and C.E.D. provided vital conceptual insights for study design, assisted with study subject accrual and data collection, and prepared figures; and all authors reviewed and edited the manuscript.
Conflict-of-interest disclosure: S.J., B.L., K.Z., and S.M.D. have a US provisional patent application pending. S.J. is supported by research and innovation grants for pediatric genomics from the Cincinnati Children's Hospital Medical Center. B.L. is supported by an American Society for Blood and Marrow Transplantation New Investigator Award. The remaining authors declare no competing financial interests.
Correspondence: Sonata Jodele, Division of Bone Marrow Transplantation and Immune Deficiency, Cincinnati Children’s Hospital Medical Center, 3333 Burnet Ave, MLC 11027, Cincinnati, OH 45229; e-mail: sonata.jodele@cchmc.org.

![Figure 4. TMA and race effect on survival among patients who received a transplant. Kaplan-Meier survival curves (total n = 333). HSCT survival is inferior in (A) HSCT patients who developed TMA (63% vs 85% 1-year overall survival [OS] and 52% vs 83% 3-year OS; P = .0001) and in (B) nonwhite HSCT recipients (56% vs 81% 1-year OS and 56% vs 75% 3-year OS; P = .0009). (C-D) Effect of TMA and race combined on HSCT survival. (C) Race did not have significant influence on survival in HSCT recipients without TMA (73% vs 87% 1-year OS and 73% vs 84% 3-year OS; P = .12). (D) Nonwhite HSCT recipients who developed TMA had significantly inferior survival compared with whites who developed TMA (37% vs 69% 1-year OS and 37% vs 55% 3-year OS, P = .04).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/8/10.1182_blood-2015-08-663435/4/m_989f4.jpeg?Expires=1765959500&Signature=MSSqOfYH9sp81se2D6w~yZtFIyxH8d0MkoiBdnDCvMGQyCYb60Rg0Q7f7gno6njeG3wlw~T3f5uTkt7NfFJD0aHp17jm4F3Mld9HALzI~4VmTLAtF4gWTyPIA3-X6Q8g71bnQcM81vw-hSU1YGttmoMJrvt3Fei2DtyOmJmoKuN9Q3KGwBWmWB~IsR3xPP3iXo0XApdMBd77kIwApW3YKZSR3mZxPlrp0MSwKoJfrJlwzfYw1CZfGrxgovV532FyYXhVoS~5GTWHxDpYw13vJelgwhapF836dUoc9v3H5uT0w1bopwfi68uW60LYsB~XLbBpehhmV1WOnhwN2EVSVA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
