Key Points
A phenotype with albinism, early-onset seizures, neurodevelopmental delay, infection susceptibility, and neutropenia is caused by AP3D1 mutations.
AP3δ deficiency destabilizes the AP3 complex and defines a novel type of Hermansky-Pudlak syndrome with severe neurologic involvement.
Abstract
Genetic disorders affecting biogenesis and transport of lysosome-related organelles are heterogeneous diseases frequently associated with albinism. We studied a patient with albinism, neutropenia, immunodeficiency, neurodevelopmental delay, generalized seizures, and impaired hearing but with no mutation in genes so far associated with albinism and immunodeficiency. Whole exome sequencing identified a homozygous mutation in AP3D1 that leads to destabilization of the adaptor protein 3 (AP3) complex. AP3 complex formation and the degranulation defect in patient T cells were restored by retroviral reconstitution. A previously described hypopigmented mouse mutant with an Ap3d1 null mutation (mocha strain) shares the neurologic phenotype with our patient and shows a platelet storage pool deficiency characteristic of Hermansky-Pudlak syndrome (HPS) that was not studied in our patient because of a lack of bleeding. HPS2 caused by mutations in AP3B1A leads to a highly overlapping phenotype without the neurologic symptoms. The AP3 complex exists in a ubiquitous and a neuronal form. AP3D1 codes for the AP3δ subunit of the complex, which is essential for both forms. In contrast, the AP3β3A subunit, affected in HPS2 patients, is substituted by AP3β3B in the neuron-specific heterotetramer. AP3δ deficiency thus causes a severe neurologic disorder with immunodeficiency and albinism that we propose to classify as HPS10.
Introduction
Syndromes of albinism associated with immunodeficiency include Chediak-Higashi Syndrome (CHS), Griscelli Syndrome type 2 (GS2), Hermansky-Pudlack Syndrome type 2 (HPS2), HPS9, and endosomal-adaptor protein p14 deficiency.1 The immunodeficiency is characterized by variable infection susceptibility associated with impaired lymphocyte degranulation, cytotoxicity, and neutropenia. The cytotoxicity defect is linked to a risk of developing hemophagocytic lymphohistiocytosis (HLH) mainly in CHS, GS2, and HPS2.2-4 Chronic neutropenia is characteristic in HPS25 and late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 deficiency,6 whereas it is usually transient in CHS and GS2. Apart from immunodeficiency, albinism syndromes can also present with other systemic manifestations, including bleeding as a result of a storage pool disorder (HPS2, CHS)7 and progressive lung fibrosis (HPS2)8 as well as neurologic disorders (CHS, GS2).9,10
The pathophysiological basis of these partly overlapping syndromes is a disturbed biogenesis and transport of secretory lysosome-related organelles.11,12 These secretory organelles are found in several cell types for secretion of specific, cell-type-dependent proteins and peptides. They are important for diverse physiological processes, including pigmentation of eyes, hair, and skin13 ; release of small molecules by δ granules from platelets at the site of blood vessel damage that facilitate platelet adhesion and activation7 ; secretion of perforin and granzyme by lytic granules in cytotoxic lymphocytes11,14 ; and secretion of surfactant by lysosome-related lamellar bodies in pulmonary alveolar type II cells.15 A distinct, but in its protein components, partly overlapping molecular machinery is responsible for the biogenesis and transport of secretory lysosome-related organelles in different cell types. This explains the variable combination of different clinical manifestations in genetic diseases that interfere with this pathway.
A key protein complex involved in lysosome biogenesis is adaptor protein 3 (AP3) complex. This adaptor protein complex is involved in the budding of vesicles from early endosomal compartments and is also involved in the correct sorting of integral membrane proteins from the endosome to the secretory lysosome.16,17 This includes sorting of LAMP-1 and LAMP-2 to secretory lysosomes,18 of tyrosinase to melanosomes,13 and of neutrophil elastase to neutrophil granules.19 In the absence of AP3, missorting of these proteins to the cell membrane has been observed. Two AP3 heterotetramers can be distinguished: the ubiquitous AP3 consists of two large subunits, β3A and δ, an intermediate subunit µ3A, and a small subunit σ3, which can be one of two isoforms, A or B.20 A second form of AP3 is specific for the central nervous system and consists of β3B, µ3B, one of the two σ3 isoforms, and the shared δ subunit.21 Deficiency of the AP3β3A subunit, which is encoded by AP3B1, results in HPS2,5 whereas mutations in the other subunits of the AP3 complex have so far not been associated with human disease.
Here we describe a patient with albinism and immunodeficiency with some characteristic features of HPS2 and additional neurologic symptoms from birth, including microcephaly, severe neurodevelopmental delay, generalized seizures, and hearing impairment. An overt tendency for bleeding was not observed. We detected a homozygous frameshift deletion in AP3D1. Reconstitution of patient T cells with wild-type AP3D1 corrected the degranulation defect. In mocha mice, Ap3d1 mutations cause an HPS-like phenotype with severe seizures and hearing loss.22 Thus, AP3δ deficiency causes a new severe neurologic disorder with immunodeficiency and albinism, which we propose to classify as HPS-10.
Methods
Informed consent
Informed consent was obtained from the patients’ family according to the guidelines of the local ethics committee (Ethics Committee, University of Freiburg, approval number 192/08).
Genetic analysis
For exome sequencing, 3 μg of DNA was fragmented by using sonication technology (Diagenode, Seraing, Belgium). The fragments were end repaired and adaptor ligated. After size selection, the library was subjected to the enrichment process by using the Nimblegen SeqCap EZ Human Exome Library v2.0 (Roche, Madison, WI) enrichment kit and sequenced by using the Illumina HiSeq2000 instrument (Illumina, San Diego, CA). Data analysis of filter-passed reads was done with Burrows-Wheeler Aligner-short in combination with Genome Analysis Toolkit and SAMtools as implemented in Varbank (Cologne Center for Genomics). In-house scripts were applied for filtering against dbSNP, the 1000 Genomes Project, and our database of exome variants. We focused on rare missense, nonsense, frameshift, and splice site mutations assuming autosomal recessive inheritance. Further criteria for variant selection were coverage of more than 6 reads, minimal quality score of 10, and minor allele frequency <1%.
Selected variants were confirmed by conventional sequencing. Respective exons were amplified by polymerase chain reaction, and products were directly sequenced by using the BigDye Terminator v.1.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA) on a 3730 DNA Analyzer (Applied Biosystems). Primer sequences are available upon request.
Protein analysis
Lysates were prepared from magnetically activated cell sorter–purified CD8 T cells or virally reconstituted phytohemagglutinin/interleukin-2 (PHA/IL-2) blasts and sorted for green fluorescence protein (GFP) expression by using NP-40 (Igepal, Sigma) lysate buffer with protease inhibitor cocktail (Roche). Lysates were diluted with 1× sample buffer (Novex NuPAGE lithium dodecyl sulfate Sample Buffer [4×]) and 1× reduction agent (Novex NuPAGE Sample Reducing Agent [10×]). Commercial gels were used (Novex NuPAGE 4% to 12% Bis-Tris Protein Gels [1.0 mm]) with 1× 0.1% sodium dodecyl sulfate (SDS) running buffer (NuPAGE 2-(N-morpholino)ethanesulfonic acid SDS Running Buffer [20×]). Protein transfer was performed on nitrocellulose membrane overnight with constant 220 mA and 1× running buffer plus 10% methanol and 0.1% SDS (NuPAGE Transfer Buffer [20×]). Membranes were blocked with 5% milk in 1× phosphate-buffered saline (PBS) with 0.05% Tween-20 (PBS-T) for 1 hour. Antibodies specific for AP3δ (rabbit-Dinah [1:200] or delta2 [1:500]), AP3β (rabbit-Marlene [1:100]), AP3µ (rabbit-Diane [1:500]), or AP3σ (rabbit-Buttercup [1:500]) in 1% milk in 1× PBS-T were incubated overnight at 4°C. All antibodies were provided by M.S. Robinson and A. Peden and raised against fusion proteins encoding the AP3δ subunit.23,24 Secondary anti-rabbit immunoglobulin G (IgG) antibody (Jackson) was incubated in 5% milk with 1× PBS-T for 30 minutes.
Degranulation and cytotoxicity assays
Natural killer (NK) –cell assays were performed as described previously.25 Degranulation of cultured transduced T cells was analyzed after resting cells for 24 hours in medium without IL-2. T cells were stimulated with 3 µg/mL plate-bound anti-CD3 (OKT3, eBioscience) or left in the medium. Anti-CD107a phycoerythrin (BD Pharmingen) was added for the 3-hour stimulation time and was again used in combination with anti-CD8 allophycocyanin (BD Pharmingen) for surface staining. Cells were analyzed with flow cytometry (Navios flow cytometer, Beckman Coulter) and FlowJo software.
Functional reconstitution of T cells
For generating an AP3D1-encoding retrovirus, Phoenix-Ampho cells were transfected with the wild-type AP3D1 gene by using PromoFectin (PromoKine) with the packaging vector pML and pMX-AP3δ1-IRES-GFP. Viral supernatant was harvested after 48 hours of culture and used for viral transduction. T-cell cultures were prepared for transduction by stimulating peripheral blood mononuclear cells (PBMCs) with PHA (1.25 μg/mL; Remel) and ∼100 U/mL human IL-2 (produced from a transfected cell line) in the presence of irradiated (30 Gy for 5 minutes) allogeneic PBMCs as feeder cells. Cells were cultured in Iscove modified Dulbecco medium (Gibco) with 10% human serum (Sigma) and 1% penicillin and streptomycin (Gibco). Cells were restimulated with fresh IL-2 and feeder cells every 14 to 18 days. Two days after restimulation, cells were resuspended in viral supernatant (containing pMX-AP3δ1-IRES-GFP or pMX-IRES-GFP as a negative control) and plated in a 24-well nonculture tissue plate coated with retronectin (10 μg/mL; TAKARA BIO), followed by 2 hours of centrifugation at 32°C. T-cell cultures were further propagated by repeated restimulation until the day of analysis.
Immunofluorescence staining
PHA/IL-2 blasts (5 × 104 cells per spot) from the patient reconstituted with either pMX-AP3δ1-IRES-GFP or pMX-IRES-GFP were adhered to Hendley-Essex multispot microscopy slides in serum-free medium (Dulbecco’s modified Eagle medium; Gibco) for 25 minutes at 37°C and fixed with 3% paraformaldehyde for 15 minutes at room temperature. Cells were blocked for 1 hour in PBS, 1% bovine serum albumin, and 0.4% Saponin in 1× PBS and were incubated with anti-AP3δ (mouse-SA4, provided by A. Peden18 ) in blocking buffer for 1 hour at room temperature, followed by extensive washes and incubation with secondary antibody (Alexa Fluor 568 Goat Anti-Mouse IgG; Invitrogen, A-11031) for 35 minutes at room temperature. Nuclei were stained with Hoechst in 1× PBS for 10 minutes at room temperature. Samples were washed before being mounted with nonsetting mounting medium (Vectashield; Vectalabs) and were imaged by using an Andor Revolution spinning disk system with CSU-X1 spinning disk (Yokogawa) with Borealis system at 1024 × 1024 pixels, pixel size 13 × 13 μm camera (iXon Ultra DU888, Andor) IX81 microscope (Olympus) with the 100× objective (numerical aperture 1.45) and 2.0× magnification lenses. Laser excitation was at 405, 488, and 561 nm. Images were acquired by using IQ3 cell imaging software (Andor) and were processed by using Imaris software (Bitplane).
Results
Patient
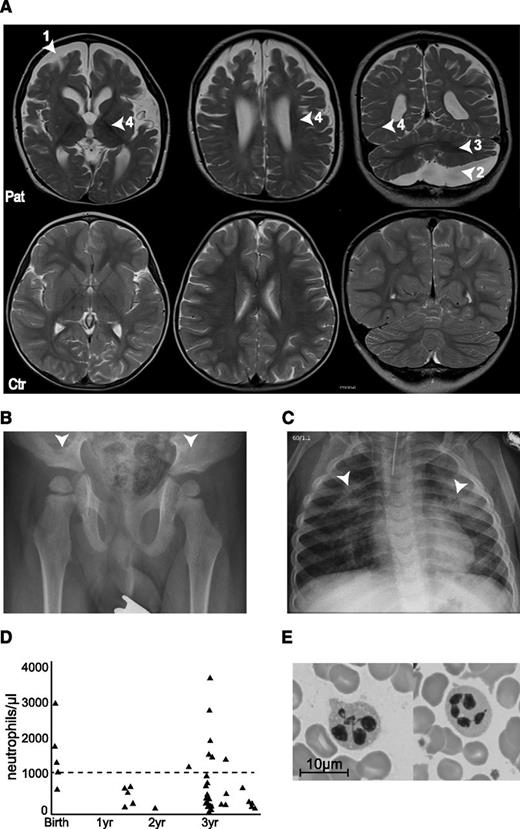
The index patient had early-onset seizures, severe developmental delay, dysmorphic features, and infection susceptibility associated with neutropenia; he was the first child of consanguineous Turkish parents (first cousins). The mother had secondarily generalized focal epilepsy that was treated until she was age 18 years and the father was healthy. A younger brother was affected by osteopetrosis caused by a homozygous TCIRG1 (OC116) mutation. At birth, the patient was microcephalic (length, 50th percentile; head circumference, 3rd percentile) with large, low set ears, retrognathia with a Pierre-Robin sequence, hypotelorism, and a flat philtrum. At age 1 week, he developed recurrent apneas followed by a focal seizure at age 11 days and increasingly dystonic movement patterns with truncal hypotonia. Within the first years of life, he developed spasms, tonic and myoclonic focal seizures, and generalized tonic-clonic seizures that occurred up to 30 times daily and were poorly controlled with multiple antiepileptic drug regimens. The electroencephalogram showed multifocal spike slow wave activity and no physiological differentiation. A hearing test revealed bilaterally reduced otoacoustic potentials and a reduced brainstem-evoked response audiometry. He made little developmental progress before his death at age 3.5 years and maintained general hypotonia, lack of head control with little spontaneous movement, and no ocular fixation. He had significant feeding difficulties. Oculocutaneous albinism was associated with poorly pigmented hair and intermittent nystagmus. Brain magnetic resonance imaging at age 3.5 years showed atrophy of the telencephalon and enlarged external and internal cerebrospinal fluid spaces as well as a large arachnoidal cyst in the posterior fossa. Furthermore, the myelination was insufficient for his age corresponding rather to that of a 15-month-old child. Cerebellar white matter was hypointense, which indicated myelin only projection fibers, and central parts of the supratentorial white matter were myelinated involving some subcortical domains but not U-fibers (Figure 1A). A hip radiograph showed flat acetabulae (Figure 1B).
Clinical phenotype of the patient. (A) Magnetic resonance imaging (MRI) scans of the brain at 3.5 years (upper row), with arrowheads indicating (1) atrophy of the telencephalon, (2) arachnoidal cyst in the posterior fossa, (3) insufficient myelination, and (4) enlarged external and internal cerebrospinal fluid spaces. The lower row shows MRI scans of a healthy 3.5-year-old child. (B) X-ray of the pelvis showing flat, dyplastic acetabulae. (C) X-rays of the chest showing chronic interstitial pneumonia. (D) Neutrophil counts over time. Dashed line at 1000 neutrophiles/µL represents the clinical limit for neutropenia. (E) Bone marrow smear showing hypersegmented neutrophils.
Clinical phenotype of the patient. (A) Magnetic resonance imaging (MRI) scans of the brain at 3.5 years (upper row), with arrowheads indicating (1) atrophy of the telencephalon, (2) arachnoidal cyst in the posterior fossa, (3) insufficient myelination, and (4) enlarged external and internal cerebrospinal fluid spaces. The lower row shows MRI scans of a healthy 3.5-year-old child. (B) X-ray of the pelvis showing flat, dyplastic acetabulae. (C) X-rays of the chest showing chronic interstitial pneumonia. (D) Neutrophil counts over time. Dashed line at 1000 neutrophiles/µL represents the clinical limit for neutropenia. (E) Bone marrow smear showing hypersegmented neutrophils.
From age 8 months, he developed recurrent bronchitis and pneumonia. Chest x-rays revealed chronic interstitial pneumonia (Figure 1C). He had persistent hepatosplenomegaly and febrile episodes that were frequently associated with severe neutropenia (Figure 1D) but did not develop thrombocytopenia or anemia. Bone marrow showed moderate dysplasias (Figure 1E) with full maturation of all lineages. He had asymptomatic seroconversion to Epstein-Barr virus and cytomegalovirus. Abnormal bleeding was not noted. The patient died at the age of 3.5 years as a result of septic pneumonia.
Immunologic investigation and exclusion of HLH
In the context of his infection susceptibility and hepatosplenomegaly associated with albinism, he received an immunologic assessment. He had normal immunoglobulin levels and vaccine responses, and IgE was elevated (3990 U/mL). Lymphocyte counts were normal with an inverted CD4/CD8 ratio (0.53) during episodes of pneumonia. Because several immunodeficiencies with albinism are associated with impaired lymphocyte cytotoxicity, we analyzed expression of the degranulation marker CD107a on fresh (Figure 2A) and IL-2–stimulated NK cells (Figure 2B) after incubation with the NK-sensitive target cell K562. NK-cell degranulation was clearly abnormal. These results were confirmed in a 51Cr release assay (Figure 2C). T-cell degranulation was impaired but not absent (Figure 2D). A bone marrow aspiration performed in 1 of the febrile neutropenic episodes showed no evidence of hemophagocytosis. Fibrinogen, triglycerides, and ferritin were normal and sCD25 was below 2400 U/mL. Criteria for HLH26 were not fulfilled.
Impaired degranulation and cytotoxicity of patient NK cells. (A) Ex vivo degranulation of NK cells (CD3–CD56+) from the patient (Pat) and a healthy donor control (Ctr) after incubation with medium (left panel) or with K562 cells (middle panel) as assessed by flow cytometric analysis of CD107a surface expression. The right panel shows the difference in CD107a expression between unstimulated and stimulated cells from the patient (solid circle) and a day control (open square) relative to healthy controls (gray shaded area). The dashed line represents the level below which NK-cell degranulation has the best positive and negative predictive value for a mutation in a gene relevant for cytotoxicity in an unfiltered cohort of patients with HLH. (B) Degranulation of NK cells prestimulated with IL-2 and PHA for 48 hours. (C) Cytotoxicity of patient and control PBMCs on K562 target cells without (black symbols) or after overnight pre-incubation with IL-2 (gray symbols). (D) T-cell degranulation. Cultured T cells from the patient and a healthy control were incubated in medium or stimulated with 3 µg/mL of plate-bound anti-CD3. The increase in CD107a expression upon stimulation is shown as an overlay of histograms of unstimulated (dotted line) and stimulated (solid line) cells.
Impaired degranulation and cytotoxicity of patient NK cells. (A) Ex vivo degranulation of NK cells (CD3–CD56+) from the patient (Pat) and a healthy donor control (Ctr) after incubation with medium (left panel) or with K562 cells (middle panel) as assessed by flow cytometric analysis of CD107a surface expression. The right panel shows the difference in CD107a expression between unstimulated and stimulated cells from the patient (solid circle) and a day control (open square) relative to healthy controls (gray shaded area). The dashed line represents the level below which NK-cell degranulation has the best positive and negative predictive value for a mutation in a gene relevant for cytotoxicity in an unfiltered cohort of patients with HLH. (B) Degranulation of NK cells prestimulated with IL-2 and PHA for 48 hours. (C) Cytotoxicity of patient and control PBMCs on K562 target cells without (black symbols) or after overnight pre-incubation with IL-2 (gray symbols). (D) T-cell degranulation. Cultured T cells from the patient and a healthy control were incubated in medium or stimulated with 3 µg/mL of plate-bound anti-CD3. The increase in CD107a expression upon stimulation is shown as an overlay of histograms of unstimulated (dotted line) and stimulated (solid line) cells.
Unknown homozygous mutation in gene coding for the δ subunit of the AP3 complex
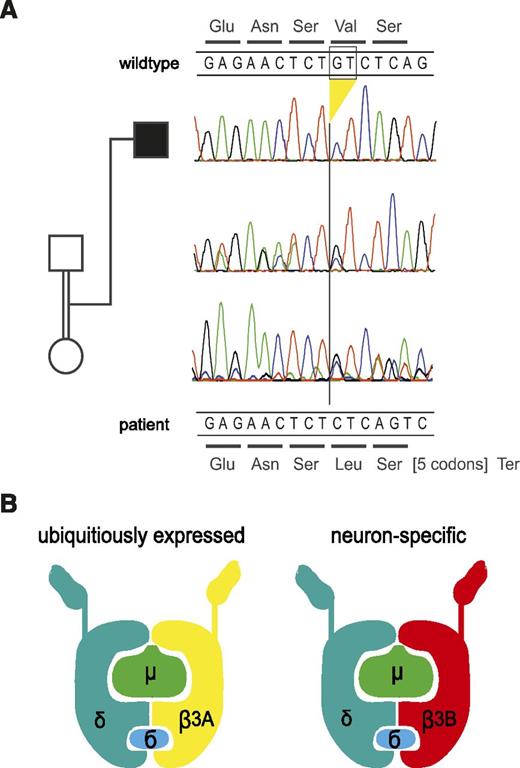
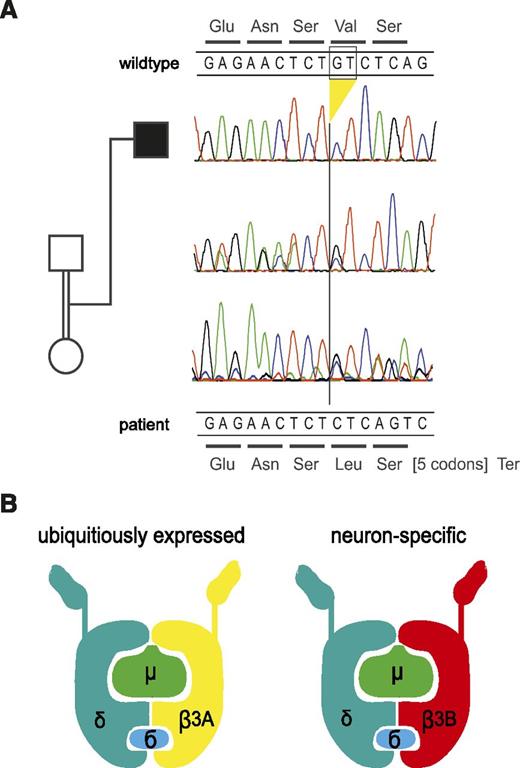
The clinical and immunologic findings suggested a genetic disorder of lysosomal trafficking associated with oculocutaneous albinism and immunodeficiency. The combination of facial and acetabular dysplasia, neutropenia, and interstitial lung disease was most consistent with HPS2, which is caused by mutations in AP3B1,5 the gene encoding AP3β3A. However, we did not find mutations in the gene. There were also no mutations in the LYST (mutated in CHS),27 RAB27A (GS2),28 and LAMTOR2 (p14 deficiency) genes.6 We then performed whole exome sequencing by using DNA extracted from peripheral blood. The patient’s exome was covered with a mean depth of 103 reads, 97% of the exons were covered >10× and 95% were covered >20×. In all, 84 M unique reads were aligned to the reference human genome, and 78 M reads were uniquely mapped to target sequences. Filtering revealed 29 homozygous autosomal variants. Assuming homozygosity by descent, we further concentrated on larger homozygous intervals, which covered 440 Mb in total. Nineteen variants resided in these regions, including 17 missense variants, 1 potential splicing variant in AOC1, and 1 frameshift deletion. Among these variants, the mutations in AP3D1 had the most plausible relationship to the clinical phenotype, especially in synopsis with the AP3d1 mouse mutant and its HPS-like phenotype (supplemental Table 1, available on the Blood Web site). The deletion of GT at positions c.3565-3566 in exon 32 of AP3D1 (c.3565_3566delGT; NM_001261826; Figure 3A) leads to a frameshift and a termination codon after 7 residues (p.Val1189Leufs*8) (Figure 3A). The deletion was not identified in the 1000 Genomes Project or the National Heart, Lung, and Blood Institute Exome Sequencing Project. The mutation affects both expressed isoforms of AP3D1; in isoform 2, which is the shorter splicing variant lacking exons 23 and 24, the mutation is found in exon 30 (c.3379_3380delGT; NM_003938). AP3D1 is expressed not only in the ubiquitous AP3 complex but also in the neuronal form (Figure 3B).
Genetic analysis and model of the AP3 complex. (A) Electropherograms of the section of exon 32 harboring the homozygous deletion in the patient. The deletion leads to a frameshift at codon 1189 and a termination codon after 7 residues. The encoded wild-type peptide sequence is shown at the top, and the codons of the mutant sequence are shown below the DNA sequences. The parents are heterozygous for the deletion as expected. For technical reasons, the reverse strand was sequenced in the DNA sample of the father but the complementary sequence is depicted here (ie, it is shown in the same direction as the other samples). (B) Model of the AP3 protein complex in the ubiquitous and the neuronal form (adapted from Odorizzi et al31 ). AP3δ is an essential part of both complexes, whereas AP3β3A, mutated in HPS2, is substituted by AP3β3B in neuronal cells.
Genetic analysis and model of the AP3 complex. (A) Electropherograms of the section of exon 32 harboring the homozygous deletion in the patient. The deletion leads to a frameshift at codon 1189 and a termination codon after 7 residues. The encoded wild-type peptide sequence is shown at the top, and the codons of the mutant sequence are shown below the DNA sequences. The parents are heterozygous for the deletion as expected. For technical reasons, the reverse strand was sequenced in the DNA sample of the father but the complementary sequence is depicted here (ie, it is shown in the same direction as the other samples). (B) Model of the AP3 protein complex in the ubiquitous and the neuronal form (adapted from Odorizzi et al31 ). AP3δ is an essential part of both complexes, whereas AP3β3A, mutated in HPS2, is substituted by AP3β3B in neuronal cells.
The mutation allows strongly reduced AP3δ protein expression and impairs stable formation of the AP3 complex
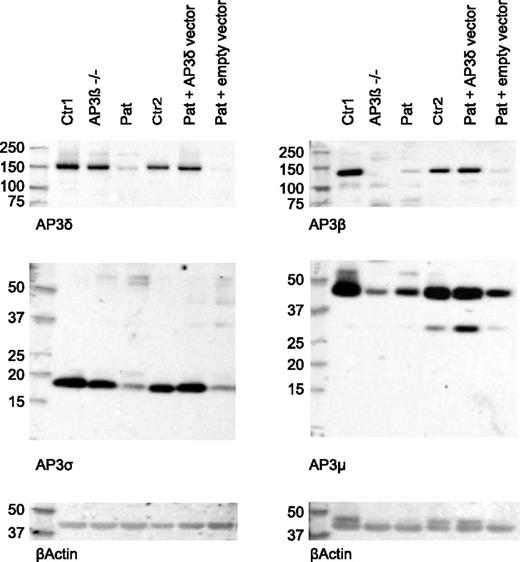
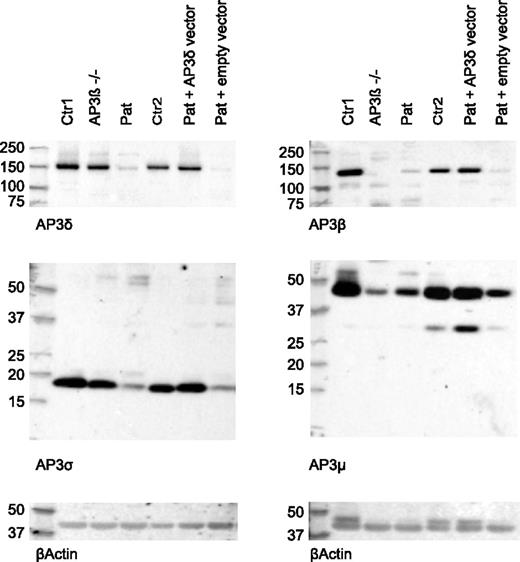
Next, we analyzed the presence of the 4 subunits of the AP3 complex in patient CD8 T cells by western blot analysis. AP3δ protein was significantly decreased compared with cells from a healthy control and an AP3β3A-deficient HPS2 patient. Consistent with an unstable heterotetramer formation, protein levels of other subunits (AP3β3A, AP3σ, and AP3µ) of the AP3 complex were clearly reduced (Figure 4). Consistent with published data,24 the control cells from the HPS2 patient showed only reduced levels of AP3β3A and AP3µ, whereas AP3δ and AP3σ were present at normal levels. Note that the molecular sizes of the 4 AP3 subunits observed in these experiments showed slight differences compared with the sizes originally reported,23 although the same antibodies were used. We ascribe these differences to the use of different gel systems and markers.
Reduced expression of AP3δ affects the stability of the AP3 complex. Western blot analysis of lysates from magnetically activated cell sorter (MACS) –purified CD8 T cells of 2 healthy controls: an AP3β3A-deficient patient and the index patient (lanes 1-4). Two separate blots are shown that used rabbit antibodies against AP3δ and AP3σ (left) and AP3β and AP3μ (right). β-actin was used as a loading control. AP3δ protein was analyzed by using 2 different antibodies23,24 in 3 independent experiments with similar results. Lanes 5 and 6 show blots from patient PHA/IL-2 blasts retrovirally transduced with an AP3D1-expressing or empty vector analyzed in the same experiment. These cells were sorted by using fluorescence-activated cell sorting (FACS) for GFP-positive cells to enrich for successfully transduced cells.
Reduced expression of AP3δ affects the stability of the AP3 complex. Western blot analysis of lysates from magnetically activated cell sorter (MACS) –purified CD8 T cells of 2 healthy controls: an AP3β3A-deficient patient and the index patient (lanes 1-4). Two separate blots are shown that used rabbit antibodies against AP3δ and AP3σ (left) and AP3β and AP3μ (right). β-actin was used as a loading control. AP3δ protein was analyzed by using 2 different antibodies23,24 in 3 independent experiments with similar results. Lanes 5 and 6 show blots from patient PHA/IL-2 blasts retrovirally transduced with an AP3D1-expressing or empty vector analyzed in the same experiment. These cells were sorted by using fluorescence-activated cell sorting (FACS) for GFP-positive cells to enrich for successfully transduced cells.
Retroviral reconstitution of primary T cells restores AP3 complex formation and T-cell degranulation
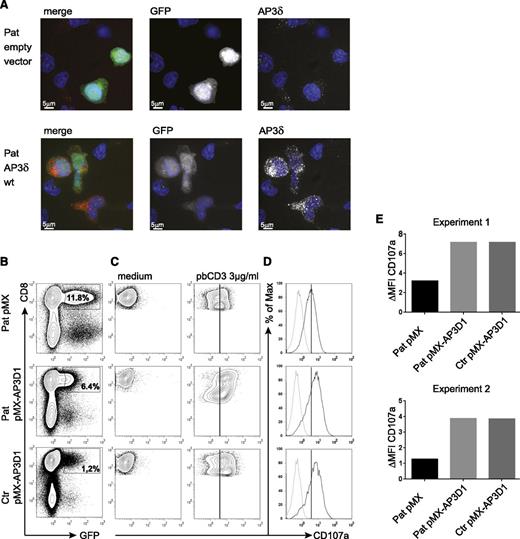
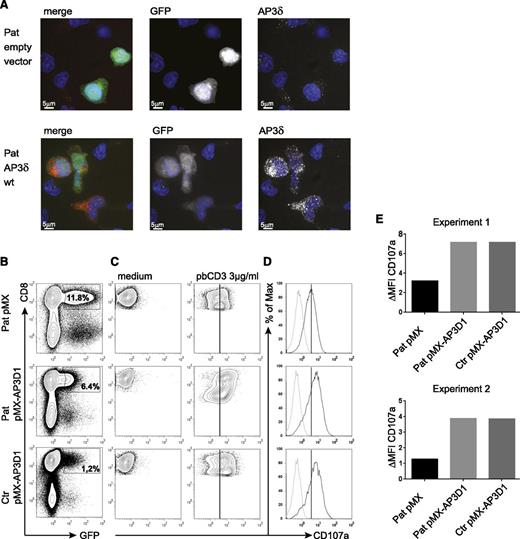
To address the question of whether the mutant AP3δ is directly responsible for the destabilization of the AP3 complex and the impaired lysosomal trafficking in patients’ cells, we generated a T-cell culture from patients’ PBMCs, which was maintained with PHA, IL-2, and feeder cells. The T cells were transduced with a retrovirus expressing either a pMX-AP3D1-IRES-GFP construct with wild-type AP3D1 or pMX-IRES-GFP as a negative control. After transduction, the culture was maintained for another 2 weeks followed by expression studies and functional tests. Successful reconstitution of GFP-positive cells with AP3D1 was demonstrated by immunofluorescence (Figure 5A), showing the expression of AP3δ as red dots in GFP-positive cells only. Furthermore, western blot analysis of reconstituted and GFP-positive sorted T cells revealed that retroviral transfection of T cells with the AP3D1 wild-type gene restored the presence of AP3δ, AP3β3A, AP3σ, and AP3µ to normal levels (Figure 4). These data confirmed that the patient mutation AP3D1 was responsible for the destabilization of the AP3 complex.
Genetic reconstitution with wild-type AP3D1 restores expression and function of the AP3 complex. (A) Immunofluorescence analysis of patient PHA/IL-2 blasts reconstituted with either empty vector or wild-type AP3D1 using mouse anti-AP3δ (SA4). Cells are shown in one 3D stack picture of 28 slices. Cells successfully transduced with the vector are GFP-positive (white in the single stains and green in the merged picture), the nucleus is stained with Hoechst (blue), and AP3δ is stained with SA4 provided by A. Peden18 (white in the single stains and red in the merged picture). The variable GFP intensity of reconstituted cells is explained by the use of an IRES-GFP construct in which AP3D1 is placed before and GFP is placed after the IRES sequence. (B) Transduced T cells were rested in medium free of IL-2 for 24 hours and then incubated in medium or stimulated with 3 µg/mL plate-bound CD3 for 3 hours. Transduction efficiencies varied between 1% and 12%. (C) CD107a expression after incubation of transduced cells with medium or plate-bound anti-CD3. Plots are gated on successfully transduced GFP-positive CD8+ cells. The solid line indicates peak CD107a expression in patient cells transduced with the empty vector. (D) Overlay of CD107a expression of transduced T cells kept in medium (dotted line) and (E) stimulated with anti-CD3 (solid line). The difference in mean fluorescence intensity (ΔMFI) of CD107a expression between cells incubated in medium vs stimulated with plate-bound anti-CD3 is shown for patient (Pat) and control (ctr) GFP-positive CD8+ T cells transduced with the indicated vector. Results are shown from 2 independent experiments.
Genetic reconstitution with wild-type AP3D1 restores expression and function of the AP3 complex. (A) Immunofluorescence analysis of patient PHA/IL-2 blasts reconstituted with either empty vector or wild-type AP3D1 using mouse anti-AP3δ (SA4). Cells are shown in one 3D stack picture of 28 slices. Cells successfully transduced with the vector are GFP-positive (white in the single stains and green in the merged picture), the nucleus is stained with Hoechst (blue), and AP3δ is stained with SA4 provided by A. Peden18 (white in the single stains and red in the merged picture). The variable GFP intensity of reconstituted cells is explained by the use of an IRES-GFP construct in which AP3D1 is placed before and GFP is placed after the IRES sequence. (B) Transduced T cells were rested in medium free of IL-2 for 24 hours and then incubated in medium or stimulated with 3 µg/mL plate-bound CD3 for 3 hours. Transduction efficiencies varied between 1% and 12%. (C) CD107a expression after incubation of transduced cells with medium or plate-bound anti-CD3. Plots are gated on successfully transduced GFP-positive CD8+ cells. The solid line indicates peak CD107a expression in patient cells transduced with the empty vector. (D) Overlay of CD107a expression of transduced T cells kept in medium (dotted line) and (E) stimulated with anti-CD3 (solid line). The difference in mean fluorescence intensity (ΔMFI) of CD107a expression between cells incubated in medium vs stimulated with plate-bound anti-CD3 is shown for patient (Pat) and control (ctr) GFP-positive CD8+ T cells transduced with the indicated vector. Results are shown from 2 independent experiments.
To test whether re-expression of the AP3 complex in patient cells was also associated with functional reconstitution, we analyzed CD8+ T-cell degranulation. Figure 5B shows the flow cytometric analysis of retrovirally transduced cells. For degranulation, we incubated the transduced T-cell cultures either with plate-bound anti-CD3 or with medium and measured the CD107a expression in successfully transduced GFP-positive CD8+ T cells (Figure 5C). As expected from previous experiments with AP3B1-deficient T cells,29 the degranulation of patient’s T cells transduced with the empty vector was reduced but not absent. Importantly, transduction with the AP3D1-containing vector fully restored the degranulation response of patient cells to levels of wild-type cells in 2 independent experiments (Figure 5D-E), whereas transduction with the empty control vector had no effect. These data support the notion that the lytic granule secretion defect results directly from the loss of AP3δ.
Discussion
Here we describe a hitherto unknown human disorder of lysosomal trafficking that shares several clinical and immunohematologic features of HPS25,29 but also manifests with severe epilepsy and neurodevelopmental delay. By using whole exome sequencing, we identified a homozygous mutation in AP3D1 in the patient. Four independent lines of evidence support our conclusion that this is the causative mutation for the phenotype.
First, the complex clinical and cellular phenotype of the described patient shares many features with that of patients with HPS2, which is caused by defects in AP3β3A,5,30 a subunit of the AP3 protein complex and an interaction partner of AP3δ.16,31 The common features include the oculocutaneous albinism, mild facial dysmorphism, microcephaly, hip dysplasia, chronic neutropenia, persistent hepatosplenomegaly, reduced cytotoxic lymphocyte degranulation, and susceptibility to airway infections. Of note, an overt bleeding disorder was not noticed, which is a characteristic feature of HPS.12,32 However, despite the unambiguous presence of a significant platelet degranulation defect, the clinical bleeding tendency is also moderate and nonspontaneous in some other forms of HPS, and its manifestation depends on the physical insult.33 Unfortunately, because we were not able to analyze platelet function in our patient ex vivo, it is unclear whether dense granule deficiency is associated with this disorder. We speculate that the patient had a platelet granule defect that was overlooked because it was a minor part of the clinical phenotype. The most striking difference with HPS2 was the neurologic phenotype that included significant neurodevelopmental delay, severe epilepsy, and a hearing disorder. Only mild neurodevelopmental delay was reported in 1 HPS2 patient.29 Neurologic involvement ranging from mild sensory deficits to mental retardation is more prominent in CHS.10
Second, the phenotype of our patient showed striking similarities to the mocha mouse strain, a naturally occurring mouse mutant with a null mutation in the Ap3d1 gene22 (Table 1). These mice have oculocutaneous albinism and reduced coat pigmentation and, importantly, they show a functional platelet defect,22 similar to AP3β3A-deficient pearl mice.34 To the best of our knowledge, neither the skeletal phenotype nor investigations on neutrophil counts or lymphocyte degranulation in mocha mice have been published. Thus, the phenotypic overlap of human and murine deficiency in the AP3β3A or AP3δ subunits of the AP3 complex remains incompletely defined. Most relevant, however, is that mocha mice show a distinct cortical excitability phenotype with brief epileptic discharges as well as balance problems resulting from otolith defects and eventual deafness.22,35 These features specifically mirror the severe neurologic phenotype observed in our patient. The analogy to the mouse model suggests that both epilepsy and deafness in our patient were causally related to the AP3δ deficiency. It should be noted that the initially described strain of mocha mice also carried grizzled (gr) and retinal degeneration 1 (Pde6brd1) alleles.36 However, the neurologic phenotype and severe hearing loss are also observed in other Ap3d1 mutant strains that do not carry these additional mutations.37
Third, we show that the identified mutation significantly reduces expression levels of the AP3δ protein. Apparently this has severe consequences for the stability of the AP3 complex because the other subunits (AP3β3A, AP3σ, and AP3µ) were also decreased in protein expression. This is in line with observations in mocha mice,22 in which analysis of brain extracts revealed that the complete absence of AP3δ expression was associated with degradation of all other subunits. Notably, the physiology of the heterotetrameric AP3 complex offers an explanation for the severe neurologic manifestations in Ap3d1 but not in Ap3b1 deficiency. The δ subunit is central to both the neuronal and the ubiquitous forms of AP3 (Figure 3B), and therefore its absence cannot be compensated. In contrast, the absence of β3A can be largely compensated in neurons but not in other tissues because of the expression of another isoform (AP3B2). The pathophysiological link from neuronal AP3 deficiency to the severe neurologic phenotype remains to be explored. On the basis of the observation of an interaction between the neuronal AP3 complex and a zinc transporter (ZnT3) in the brain, it was previously postulated that the absence of AP3 may lead to missorting of ZnT3 and result in zinc deficiency.38 In addition, in the absence of AP3, various synaptic vesicle membrane proteins are mistargeted, including synaptic vesicle chloride channel 3 (ClC-3),39 vesicular glutamate transporter 1 (VGLUT1),40 PI4KIIa,41 the vesicular GABA transporter (VGAT),42 and the synaptobrevin-1-like SNARE VAMP7.21,43 These alterations may be highly relevant for the development of the epilepsy.
Fourth, introduction of the wild-type AP3D1 gene into CD8+ T cells restored the stability of the whole AP3 complex. Although all subunits were highly reduced in unmanipulated patient cells, all of them were detected in normal levels after retroviral reconstitution with the wild-type AP3D1 gene, revealing that the patient mutation of AP3D1 was indeed responsible for the instability of the AP3 complex. Importantly, the genetic correction also led to a functional reconstitution of the granule exocytosis defect in cytotoxic lymphocytes, proving that AP3D1 deficiency is responsible for this phenotype. Overall, the impaired degranulation response of lymphocytes in our patient was similar to that of patients with HPS2.2,29 It was most evident in fresh NK cells, in which the response was very low and was indistinguishable from that of patients with familial HLH, GS2, or CHS.25,44 However, in contrast to these conditions and similar to our observations in HPS2, the response of PHA/IL-2–stimulated cytotoxic T lymphocytes was less markedly impaired. This could possibly be the result of an effect of IL-2 on the indirect vesicular transport pathway,45,46 which is independent of the AP3 complex and has a recycling function.5,18
Although these arguments provide a plausible link from AP3δ deficiency to all aspects of the patient phenotype, a contribution of other homozygous alleles (supplemental Table 1) cannot be formally excluded. The description of further patients will help to resolve this issue. Based on the preliminary definition of this complex phenotype in a single patient, what are the clinical implications of a diagnosis of AP3δ deficiency? AP3δ deficiency appears to be mainly a neurologic disorder. In our patient as well as in mocha mice,22 the severe epilepsy associated with a significant neurodevelopmental delay dominated the clinical picture. However, the immune-hematologic aspects of the disease are probably of similar importance and can be life limiting. The resulting infection susceptibility is at least in part favored by a defect in neutrophil development and, as in HPS2, probably also by functional defects in other immune cell populations.47 This includes an impaired degranulation and cytotoxicity of lymphocytes2,25,48 and could also imply impaired dendritic cell function, as demonstrated in a mouse model of HPS2.49 We previously showed that HPS2 patients carry a small but relevant risk of developing HLH.2 Asymptomatic seroconversion to Epstein-Barr virus, cytomegalovirus, and herpes simplex virus in our patient indicates that this risk is also limited in AP3δ deficiency. However, we interpret the persistent hepatosplenomegaly and recurrent fevers in our patient as subclinical HLH-associated manifestations. In the context of the poor neurologic prognosis, the hematologic manifestations may not justify hematopoietic stem cell transplantation, but monitoring neutropenia and hepatosplenomegaly should be part of the clinical management of affected patients. Monitoring should also include evaluation for chronic lung disease because the interstitial lung disease can determine long-term prognosis in some HPS variants.1,8
In summary, AP3δ deficiency further adds to the growing spectrum of human genetic disorders of biogenesis and trafficking of lysosome-related organelles and illuminates the importance of the cell type–specific synthesis of AP-complex subunits. Although we could not analyze the characteristic storage pool deficiency of HPS in our patient, we suggest classifying AP3δ deficiency as a novel type of HPS. From a clinical point of view, bleeding seems to be a minor aspect of the disease (as in HPS2), but the AP3-related pathophysiology favors classification as HPS rather than as a new congenital neurodevelopmental disease, a new variant of congenital neutropenia, or a novel primary immunodeficiency. Hence, we suggest giving the disorder the acronym HPS10, but this has to be confirmed by an appropriate taxonomy committee.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to the patient’s family for their support. The authors also thank Margaret Robinson and Andrew Peden from the Cambridge Institute for Medical Research, Cambridge, United Kingdom, for their AP3 antibodies and their interest in and discussion of this work and Ramona Schupart and Margit Rauch for excellent technical assistance.
This work was supported by grants EH 145/5-1 and SFB1160, TP1 (S.E.) and HE 3119/10-1 (H.C.H.) from the Deutsche Forschungsgemeinschaft, 01 EO 0803 (S.E.) from Bundesministerium fuer Bildung und Forschung, by the Köln Fortune Program of the Faculty of Medicine, University of Cologne (H.C.H.), by grant 100140 from the Wellcome Trust, and by the National Institute for Health Research Biomedical Research Center (G.M.G.).
Authorship
Contribution: S.A. performed protein studies and functional and reconstitution experiments and drafted the manuscript; A. Schulz, I.K.-M., and K.N. cared for the patient and provided clinical information; N.M.G.D. performed protein and microscopy studies; S.F. prepared the reconstitution vector; K.M.E. performed gene expression and protein experiments; R.P. and R.W. performed protein experiments; J.A. performed next-generation sequencing experiments; H.T. performed variant calling; P.N. supervised next-generation sequencing experiments; J.B. prepared the reconstitution vector; A. Strauss performed bone marrow analysis; H.v.B. performed the cytotoxicity assay; U.z.S. performed genetic studies; S.G. generated the T-cell line; G.M.G. supervised protein and microscopy studies and edited the manuscript; K.L. provided clinical information; H.C.H. designed the study, coordinated the project, and edited the manuscript; S.E. designed the study, coordinated the project, and drafted the manuscript; and all authors approved the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stephan Ehl, Center for Chronic Immunodeficiency, University Medical Center Freiburg, Breisacher Strasse 117, 79106 Freiburg, Germany; e-mail: stephan.ehl@uniklinik-freiburg.de; or Hans Christian Hennies, Cologne Center for Genomics, University of Cologne, Weyertal 115b, 50931 Köln, Germany; e-mail: h.hennies@uni-koeln.de.
References
Author notes
H.C.H. and S.E. contributed equally to this work.