Abstract
Objective: We applied next generation sequencing (NGS) with a panel of 206-MPN-relevant genes, in order to identify non-driver mutation profile and prognostic value of non-driver mutations in PMF and Post-PV/ET myelofibrosis.
Methods: Targeted capture sequencing assays were carried out on bone marrow DNA specimens obtained at time of referral. Gene list was shown in Figure 1. DNA libraries were prepared using Illumina standard protocol. The amplified DNA was captured by using biotinylated oligo-probes (MyGenosticsGenCap Enrichment technologies). Illumina utilizes a unique "bridged" amplification reaction that occurs on the surface of the flow cell. A flow cell containing millions of unique clusters is loaded into theHiSeq 2000 for automated cycles of extension and imaging. TheSolexa QA package, thecutadapt program, theSOAPaligner program, the Picard software, the GATK program, the Exome-assistant program, theMagicViewer, thePolyphen, the SIFT, the PANTHER and thePmut were used to bioinformatics analysis.
Results: 54 PMF patients (median age 55 [21-77]yrs; 69% males) and 17 post-PV/ET patients (median age 59[46-83]yrs; 59% males) were tested. In PMF patients, 6 (13%) subjects were categorized as DIPPS low-risk group, 25 (56%) intermediate-1-riskgroup and 14 (31%) intermediate-2-risk group. JAK2 mutations were detected in 21 subjects (47%), CALR mutations in 4 (9%) , MPL mutation in 1(2%) and Tri-negative (no detectable mutation in JAK2, CALR or MPL) in 19 (42%). In post-PV/ET MF patients, JAK2 mutations were detected in 16 (94%) and CALR mutations in 1 (6%).
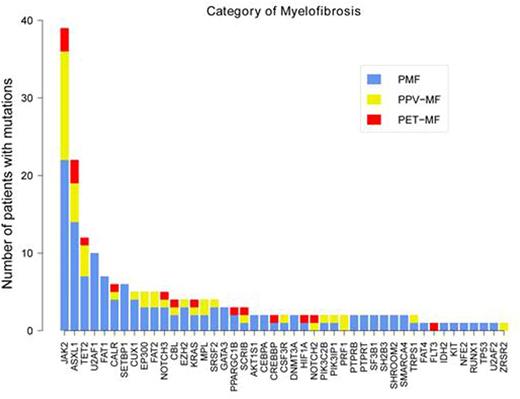
In PMF patients, mutations other than JAK2, CALR or MPL, referred as non-driver mutation, were detected in 43 (96%) patients including 89% of "Tri-negative" subjects. 12 (27%) subjects harbored one, 9 (20%) two, 12 (26.7%) three and 10 (22%) four or more. Mutational frequencies were: ASXL1 33%, U2AF1 22%, TET2 16%, FAT1 16%, SETBP1 13%, CUX1 9%, EP300 9%, SRSF2 9%, FAT 2 6.7%, NOTCH3 6.7%, EZH2 6.7%, GATA3 6.7% and <5% (at least 2 subjects) for KRAS, SF3B1, CEBPA, DNMT3A, PPARGC1B, SH2B3, PTPRB, AKTIS1, PTPRT and SHROOM2 (Figure 2).We searched for pairwise gene associations, recognizing that pairs of genes could show a tendency to eithercooccurrenceor mutually exclusivity.JAK2 shows a clear proclivity forcomutationwith TET2 (OR, 7.7, q=0.04), whereas JAK2 is mutually exclusive with CALR (OR 0.09, q=0.02).ASXL1 shows a clear proclivity forcomutationwith FAT2 (OR, 7.9, q=0.05). EP300 shows a clear proclivity forcomutationwith SRSF2 (OR, 62.2, q=0.001).
In post PV/ET MF patients, non-driver mutations were detected in 15 (88%) patients. ASXL1 was the most frequently mutated in the cohort (41%), followed by TET2 (29%) and NOTCH3 (18%) (Figure 2).Compared to patients with PMF, no splicing factor gene (U2AF1, U2AF2, SRSF2 and SF3B1) mutation was detected in patients with post-PV/ET MF.
In PMF patients, subjects classified as higher-risk (Int-2 or High risk) DIPSS group were more likely to have U2AF1 mutation (P=0.03). Patients with ASXL1 mutations were more likely to have higher WBC level (P=0.018). Patients with TET2 mutations were more likely to have higher platelet level (P=0.03), but patients with FAT1 mutations were more likely to have lower platelet level (P=0.035). In post PV/ET MF patients, subjects with ASXL1 mutations were more likely to have larger spleen size from LCM (P=0.006).
In PMF patients, follow-up data were available for 38 subjects. Date of last follow-up was June, 2016 or last contact. Median follow-up of survivors was 36 months. 8 (18%) deaths were documented. In univariate analysis, the number of non-driver mutations was considered as three categories (zero, 1-2 and more than 3). The 5-yrs PS was 100%, 81.6% and 24.4% in three categories respectively. In addition, survival was adversely affected by presence of mutant U2AF1 (P=0.009).
Conclusions: Non-driver mutations occur in about 90% of patients with PMF or Post-PV/ET MF. Mutant epigenetic genes and splicing factor genes are the most common non-driver mutations. Survival was adversely affected by the number of non-driver mutations and presence of U2AF1 mutation.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.