Abstract
Introduction
Donor cell leukemia (DCL) is a rare complication of allogeneic stem cell transplantation (allo-SCT). The leukemic transformation of otherwise healthy donor stem cells provides a useful in vivo model to study the mechanisms involved in leukemogenesis.
The objective of the present study is to study the dynamics of emergence of mutations that precede the development of DCL.
Material and methods
We report the case of a female patient diagnosed of acute lymphoblastic leukemia with t(1;19), who developed normal karyotype acute myeloid leukemia (AML) of donor origin 16 months after unrelated cord blood transplantation (UCBT).
Whole exome sequencing(SureSelect-XT Human exon capture 50Mb Agilent Technologies) was performed by next generation sequencing (Hiseq 2000 Illumina) on bone marrow samples post allo-SCT, obtained in the days +98, +189, +350, +486 (DCL diagnosis) and + 569 (DCL relapse) as well as on the UCB unit used in SCT.
The exome of bone marrow samples post allo-SCT were aligned to the human reference genome (NCBI build 37/hg19), non-synonymous variants in the coding regions of genes related to leukemia were selected and matched to the UCB exome to identify the acquired variants. Variants meeting such criteria were evaluated with three softwares (SIFT, Polyphen and Mutation Taster) to predict their functional effects. The UCB exome was aligned to the human reference genome (NCBI build 37/hg19).
Results
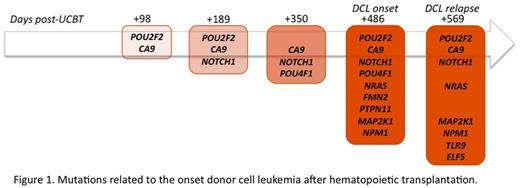
In silico variants analysis revealed progressive emergence of multiple mutations related to the development of leukemia in bone marrow samples post allo-SCT (Figure 1). Interestingly three of the mutated genes (PTPN11, NRAS, MAP2K1) are part of the same cellular pathway (RAS-MAPK).
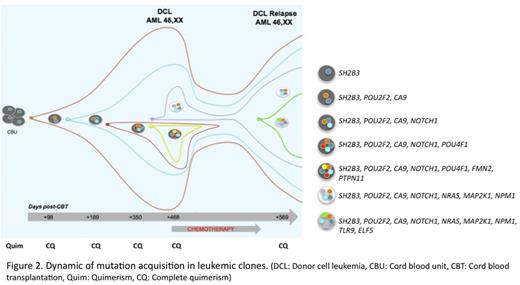
In addition, mutations in leukemic subclones that disappear after chemotherapy were indentified, as well as the acquisition of new mutations in resistant subclones (Figure 2). Finally a mutation in SH2B3 gene, a gene that encodes for a regulatory protein of the JAK-STAT pathway, which links strongly to JAK2 inhibiting the activation of the pathway, was detected in the CBU.
Conclusions
The present study reveals the acquisition of additional somatic mutations in donor cells during leukemogenesis, including mutations not previously described in AML initiation (POU2F2, CA9, POU4F1, FMN2, TLR9, ELF5).
Although the cause of DCL onset seems to be multifactorial, the infusion of a CBU with pre-leukemic potential due to mutation in SH2B3 in a context of residual toxicity in recipient as a result of pre-transplant chemotherapy, a post-transplant environment characterized by a decreased immune surveillance may well have played role in the case here reported.
The study of a greater number of DCL cases by next generation sequencing could help to understand the process of leukemogenesis.
Mutations related to the onset of donor cell leukemia after hematopoietic transplantation.
Mutations related to the onset of donor cell leukemia after hematopoietic transplantation.
Dynamic of mutation acquisitions in leukemic clones.(DCL: Donor cell leukemia, CBU: Cord blood unit, CBT: Cord blood transplantation, Quim: Quimerism, CQ: Complete quimerism)
Dynamic of mutation acquisitions in leukemic clones.(DCL: Donor cell leukemia, CBU: Cord blood unit, CBT: Cord blood transplantation, Quim: Quimerism, CQ: Complete quimerism)
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.