Abstract
Background: Acute Myeloid Leukemia (AML) without favorable and particular unfavorable cytogenetic aberrations is classified as intermediate prognosis. Recurrent somatic genomic mutations through The Cancer Genome Atlas (TCGA) have provided valuable prognostic information in AML. There are very few studies that have incorporated the cytogenetics (Cy), Next Generation sequencing (NGS) and Overall Survival (OS) in AML. Moreover prognostic NGS studies have a limited collective context without a quantifiable stratification. For the purpose of prognostic risk stratification, we utilized the TCGA data by Ley et.al and developed a system of Integer Weights for the Genomic Mutation Score (IWGMS) obtained by targeted NGS at Houston Methodist Hospital. We then correlated the IWGMS with patients' outcomes in a retrospective manner.
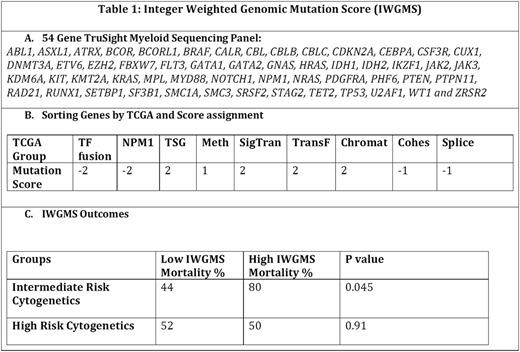
Methods: The patient data was queried from METEOR (Methodist Environment for Translational Enhancement and Outcomes Research), a clinical data warehouse that integrates existing data with internal and external research databases and national registries. We queried for the diagnosis of AML and demographics, Cy, NGS and OS. We divided the patients into 3 groups according to their MRC cytogenetic risks- Favorable (FCy), Intermediate (ICy), Poor (PCy)). Mutations in 54 genes associated with myeloid disorders were tested in NGS by using the TruSight Myeloid Sequencing Panel (Illumina) (Table 1). Briefly, genomic DNA was extracted from sorted specimens using automated methods on the MagnaPure Compact instrument (Roche Diagnostics). Sequencing libraries were prepared using TruSight reagents and sequenced on a MiSeq instrument using a 2x300 bp paired end strategy. Read alignment and variant calling for minimum of 100x amplicon coverage was performed using VariantStudio Software (Illumina). The biologic significance of detected variants was determined using VarView5 (an in-house developed bioinformatics tool). Interpretations are reviewed in a consensus conference. For the data described herein, interpretations were limited to known nonsynonymous (single nucleotide polymorphsims and insertions or deletions) somatic mutations.
Results: Of the 1200 AML patients reviewed, 100 patients meet the criteria for having the information on Cy and NGS. The mortality rate for FCy, ICy and PCy groups are 43%, 52%, and 51% respectively. IWGMS was designed by assigning a score for each genomic mutation between the range of negative 2 (-2) for good risk to positive 2 (+2) for each of 9 TCGA mutation categories (Transcription- Factor Fusion, Nucleophosmin (NPM1), Tumor Suppressor Genes, DNA-Methylation related genes, Signaling Genes, Chromatin Modifying Genes, Myeloid Transcription Factor Genes, Cohesion complex Genes and Spliceosome-complex genes). The total IWGMS for each patient was calculated by the sum of mutation scores in each 9 categories. The score greater than 3 was defined as high risk. By univariate analysis, in the ICy, high IWGMS was associated with significantly higher mortality rate than low IWGMS (80% vs 44%, p=0.045). In PCy, patients with high IWGMS and low IWGMS had similar mortality rate (50% vs 52%, p=0.910) (Table 1).
Conclusions: We have developed an IWGMS system to stratify patients with ICy into high and low risk groups. Our result demonstrated the significance of high mutation score in this group of patients, which will impact the management. The identification of genomic alterations along with the scoring system has a great potential for developing rationally-targeted therapies and improving outcomes. Further analysis along with the treatment outcomes will be presented at the ASH meeting in San Diego.
Rice:Novartis Pharma: Speakers Bureau; Alexion: Membership on an entity's Board of Directors or advisory committees, Other: Participation in registry studies; Apellis/Synergy: Other: Data Safety Monitoring Committee member; Ablynx: Other: Participation in multi-center research trial; Selexys: Other: Participation in multi-center research trial; Incyte: Other: Participation in multi-center research trial. Iyer:Genentech: Research Funding; Seattle Genetics: Research Funding; BMS: Research Funding; Incyte: Research Funding; Arog: Research Funding; Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.