Abstract
Background: Diffuse large B-cell lymphoma (DLBCL) is clinically and biologically heterogeneous. Whereas most patients are cured following conventional therapy with rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone (R-CHOP), a substantial minority are refractory or relapse. Identifying patients at high risk of poor outcomes at the time of diagnosis will make it possible to better choose alternative or experimental therapies. TCF3 (also called E2A) is a transcription factor required in B-lymphopoiesis. Gain-of-function somatic mutations that alter TCF3 or its regulatory partners are associated withBurkitt lymphoma (BL) but not DLBCL. These published findings suggest that biologically-defined, clinically-important subtypes of mature B-cell-derived lymphomas may be distinguished from one another based on differential expression of genes regulated by a master regulator of B-lymphoid biology, namely TCF3. Therefore, in the current study, we hypothesized that biologically- and clinically-important subtypes of DLBCL are distinguishable based upon differential expression of genes that are regulated in B-cells by TCF3.
Methods: Fifty-nine subjects who presented with de novo DLBCL at Kingston General Hospital from 2001 to 2010 were identified for inclusion based on the availability of clinical data and sufficient biopsy material for extraction of adequate RNA. We used publically-available microarray-based gene expression and clinical data to identify 99 candidate TCF3 target genes whose differential expression was associated with 3-year overall survival following R-CHOP therapy; these transcripts were represented in aNanoStringCodeSet. Total RNA was purified from cores harvested from representative areas of diagnostic, pre-treatment, formaldehyde-fixed, paraffin-embedded (FFPE) biopsy samples. Twenty-five transcripts whose abundance, individually, was associated with 3-year overall survival in our own cohort were chosen for further study.
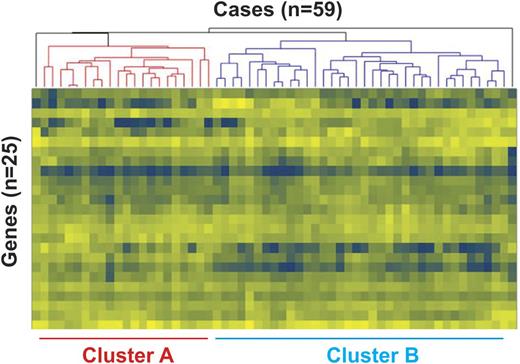
Results: Unsupervised hierarchical cluster analysis based on relative expression of the 25 TCF3 target genes resolved all except two cases into distinct clusters, denoted A and B, that contained 21 and 36 cases, respectively (heat map at left). In comparing these clusters, Cluster B was associated with rearrangement of MYC (p=0.02) as determined by FISH; no statistically significant associations were evident with age, sex, IPI score, B-symptoms, bulky disease or expression of MYC or BCL2 as determined by immunohistochemistry. Kaplan Meyer (KM) analysis revealed strikingly inferior overall and event-free survival (p=0.0005 for each) for Cluster A cases (KM curve at right).
Conclusions: Our findings demonstrate that differential expression of TCF3 target genes ascertained in pre-treatment FFPE biopsy samples of de novo DLBCL defines biologically distinctive subsets of cases with distinctive clinical outcomes following R-CHOP therapy. If validated in an independent cohort, these results may inform both clinical management and the development of new targeted therapies for high-risk cases.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.