Abstract
Background: AML is a hematological disorder resulting from proliferation and expansion of malignant myeloid cells. Expansion of malignant cells within the bone marrow and bloodstream causes severe clinical consequences. AML is a clinically challenging disease as many chemotherapeutic agents recommended in current guidelines have narrow therapeutic ranges and significant interpatient variation in treatment response. This can lead to severe therapeutic complications that are potentially fatal if concentrations are too high or failure to achieve treatment response if concentrations are too low. There is an ongoing search for predictive biomarkers to improve appropriate selection and dosing of chemotherapeutic agents. The emerging field of metabolomics has yielded promising results in the pursuit of precision medicine. However, there is still a gap in our knowledge of how metabolomics relate to treatment response to anti-leukemic agents for AML treatment. In this study, we propose to perform global metabolomics profiling of AML cell lines representing different risk groups to identify metabolic biomarkers predictive of in vitro cytotoxicity of most commonly used anti-leukemic agents.
Methods: We performed global metabolomics on eight AML cell lines (MV4.11, KG-1, HL-60, Kasumi-1, AML-193, ME1, MOLM16, THP-1) using liquid chromatography high resolution mass spectrometry (LC-HRMS). Univariate and multivariate analyses were performed on the metabolite peak intensity values from LC-HRMS using MetaboAnalyst. Each cell line was treated with varying concentrations of cytarabine. Cell viability was then determined using MTT cell survival assays. Area under the cell survival curve (AUC) and IC50 values as well as correlation between metabolites and in vitro cytotoxicity were conducted using GraphPad Prism software.
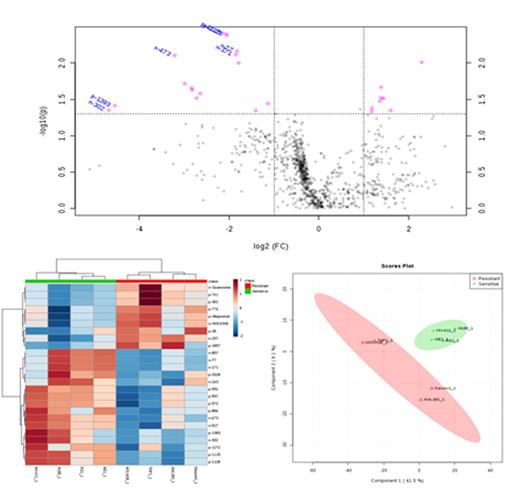
Results: A total of 2344 metabolites were detected in the global metabolome for the 6 cell lines. The metabolomes were filtered by removing metabolites present in fewer than 80% of cells or a signal below the cutoff threshold. Further analysis was conducted on the filtered dataset of 1634 metabolites. Since not all the metabolites detected have been successfully annotated, we performed an additional analysis with the dataset filtered to only include 197 annotated metabolites We classified cell lines into sensitive vs. resistant based on in vitro cytotoxicity. As shown in Figure 1, we observed significant differences in metabolic profiles (23 differentially expressed metabolites) between cytarabine sensitive vs. resistant cell lines. Multivariate analysis showed clear separation between sensitive and resistant groups, also shown in Figure 1.
Through Spearman correlation analysis, we identified 6 unique known metabolites with significant correlation with cytotoxic AUC values for cytarabine. Of these known metabolites, one also showed significant differential expression between the sensitivity groups in the filtered dataset (Guanosine). 39 currently unknown metabolites also showed significant correlation with cytotoxic AUC values for cytarabine, with 9 metabolites also showing significant differential expression between the sensitivity groups.
We expanded our correlation analysis to include an additional three commonly used chemotherapeutic agents in AML treatment regimens (clofarabine, doxorubicin and etoposide). We identified 12 unique known metabolites with significant correlation with drug sensitivity (6 for clofarabine, 6 for doxorubicin, 0 for etoposide). For unknown metabolites, 237 also showed significant correlation with cytotoxic AUC values among the four chemotherapeutic agents.
Conclusion: Overall, our results demonstrate that metabolomics differences contribute towards drug resistance. In addition, it could potentially identify predictive biomarkers for chemosensitivity to various anti-leukemic drugs. Our results provide opportunity to further explore these metabolites in patient samples for association with clinical response.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.