Abstract
Introduction: Translating the genetic and epigenetic heterogeneity underlying human cancers into therapeutic strategies represents an ongoing challenge. Large-scale sequencing efforts have identified that many hematologic malignancies, such as acute myeloid leukemia (AML), are driven by a spectrum of mutations and may require combinations of targeted agents to be treated effectively. In addition, the emergence of genetically heterogeneous subclones leading to relapse, rescue signals in the microenvironment, and tumor-intrinsic feedback pathways further necessitate combinatorial therapies. To identify combinations of targeted drugs for AML and other hematologic malignancies, we performed ex vivo profiling of pairs of small-molecule inhibitors for sensitivity against primary patient samples.
Methods: Freshly isolated primary mononuclear cells from patients (n=122) with various hematologic malignancies (AML n=58, CLL n=42, ALL n=12, and MPN or MDS/MPN n=10) were cultured in the presence of a panel of 48 drug combinations in equimolar dose series encompassing different classes of compounds, including kinase inhibitors, bromodomain inhibitors, BH3 mimetics, and histone deacetylase inhibitors. For comparison, cells were also tested against graded concentrations of each inhibitor alone, and sensitivity was assessed by MTS-based viability assay. IC50 and AUC values were derived using a probit regression model. Efficacy of each combination relative to its single agents was calculated as a Combination Ratio (CR) value, defined as the combination IC50 or AUC divided by the lowest single agent IC50 or AUC value. A CR value < 1 indicates the combination is more effective relative to the single agent. Associated clinical characteristics were obtained where possible. For the 2 largest diagnostic groups, AML and CLL, expanded panels of clinical, prognostic, mutational, cytogenetic, and surface antigen data were compiled for comparisons according to CR values for each combination.
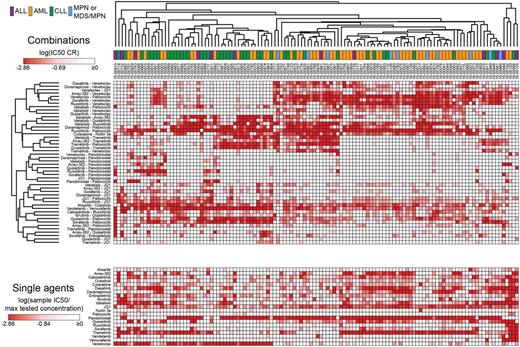
Results: Unsupervised hierarchical clustering of CR values revealed several distinct clusters (Figure 1). Myeloid leukemia patient samples were enriched within a cluster of sensitivity to combinations pairing the Bcl-2 inhibitor venetoclax with select tyrosine kinase inhibitors (dasatinib, doramapimod, sorafenib, or idelalisib). A subset of samples within this cluster showed sensitivity to combinations involving the MEK inhibitor trametinib and a second kinase inhibitor (idelalisib, palbociclib, or quizartinib). In contrast, a discrete subcluster of predominantly lymphoid leukemia patients showed sensitivity to combinations of the histone deacetylase inhibitor panobinostat in tandem with either the JAK inhibitor ruxolitinib or the multi-kinase inhibitor sorafenib. Importantly, apart from venetoclax which as a single agent demonstrated potent and selective efficacy in CLL patient samples, the single agent efficacies do not align selectively to a combination efficacy-derived cluster (Figure 1). Comparison of CR values within each of the 4 diagnostic groups revealed partial overlap in statistically significant effective combinations, while also highlighting unique sensitivities by group, such as idelalisib-quizartinib for AML and ibrutinib-quizartinib for CLL. Further relevant clinical and genetic features were compared within each of the 2 largest groups, AML and CLL. Among AML samples, patients harboring mutations in NPM1 or DNMT3A demonstrated significant sensitivity to combinations of JQ1 and sorafenib (median CR: 0.357) or JQ1 and palbociclib (median CR: 0.119), respectively. AML patients featuring surface expression of CD11b (Integrin aM) or CD58 (LFA-3) were sensitive to combinations of venetoclax and JQ1 or venetoclax and doramapimod, respectively. Among CLL samples, patients harboring deletion of 13q showed significant sensitivity to combinations of palbociclib with either venetoclax or trametinib (median CR: 0.267 and 0.116, respectively).
Conclusions: The data reveal multiple specific patterns of ex vivo drug combination efficacy beyond that of either single agent, which are associated with select, actionable diagnostic and genetic subsets, warranting their evaluation in the clinic. These findings highlight the heuristic value of an integrated approach for identifying novel treatment strategies for improved disease control and patient outcomes.
Druker:Agios: Honoraria; Ambit BioSciences: Consultancy; ARIAD: Patents & Royalties, Research Funding; Array: Patents & Royalties; AstraZeneca: Consultancy; Blueprint Medicines: Consultancy, Equity Ownership, Other: travel, accommodations, expenses ; BMS: Research Funding; CTI: Equity Ownership; Curis: Patents & Royalties; Cylene: Consultancy, Equity Ownership; D3 Oncology Solutions: Consultancy; Gilead Sciences: Consultancy, Other: travel, accommodations, expenses ; Lorus: Consultancy, Equity Ownership; MolecularMD: Consultancy, Equity Ownership, Patents & Royalties; Novartis: Research Funding; Oncotide Pharmaceuticals: Research Funding; Pfizer: Patents & Royalties; Roche: Consultancy. Tyner:Constellation Pharmaceuticals: Research Funding; Agios Pharmaceuticals: Research Funding; Takeda Pharmaceuticals: Research Funding; Inctye: Research Funding; Genentech: Research Funding; Aptose Biosciences: Research Funding; Seattle Genetics: Research Funding; Array Biopharma: Research Funding; AstraZeneca: Research Funding; Leap Oncology: Consultancy; Janssen Research & Development: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.