In this issue of Blood, Yan et al report their findings in natural killer/T-cell lymphoma (NKTL) and present experimental evidence that EZH2 can act as a transcriptional activator, independent of its methyltransferase activity, when phosphorylated by JAK3. This may open up the prospect of new treatment options for a variety of neoplasms, where EZH2 is known to be involved.1
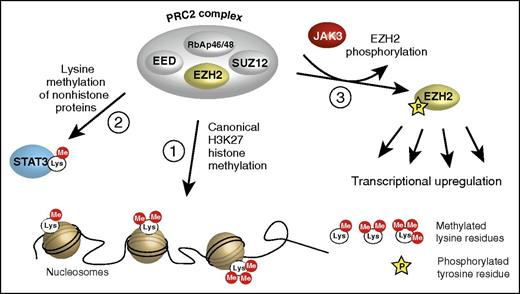
Three ways of EZH2 action. (1) The “canonical” action as a transcriptional repressor via methylation of lysine 27 in histone 3 (H3K27); only core components of the PRC2 complex are shown. (2) The transcriptional activation of nonhistone proteins by lysine methylation. (3) The phosphorylation by JAK3 turning EZH2 into a transcriptional activator, described by Yan et al. EED, embryonic ectoderm development; RbAp46/48, retinoblastoma-associated proteins 46/48; SUZ12, suppressor of zeste 12.
Three ways of EZH2 action. (1) The “canonical” action as a transcriptional repressor via methylation of lysine 27 in histone 3 (H3K27); only core components of the PRC2 complex are shown. (2) The transcriptional activation of nonhistone proteins by lysine methylation. (3) The phosphorylation by JAK3 turning EZH2 into a transcriptional activator, described by Yan et al. EED, embryonic ectoderm development; RbAp46/48, retinoblastoma-associated proteins 46/48; SUZ12, suppressor of zeste 12.
EZH2 (polycomb repressive complex 2 [PRC2] subunit) is the central core component of the PRC2, a multiprotein complex that methylates lysine 27 of histone 3 (H3K27) and acts as a transcriptional repressor by silencing expression of various target genes (see figure, path 1). The PRC2 complex is involved in a number of important physiological processes, including X-chromosome inactivation, stem cell maintenance, lymphopoiesis, cell adhesion, and migration among others.2,3 Besides its “canonical” function as PRC2-dependent H3K27 methylator, EZH2 is also capable of methylating a number of nonhistone proteins, such as STAT3, thus acting as a transcriptional activator4 (see figure, path 2).
EZH2 has been found to be mutated in various hematopoietic neoplasms, primarily germinal center B-cell–like diffuse large B-cell lymphomas and follicular lymphomas, myelodysplastic syndromes (MDSs), and myeloproliferative neoplasms (MPNs). Two different types of mutations have been observed: loss-of-function (inactivating) and gain-of-function (activating) mutations. Inactivating mutations were reported not only from MDS and MPN patients but also from those with acute T-cell leukemia.5,6 Activating heterozygous mutations which are mostly found in lymphomas predominantly affect the tyrosine residue 641 (Y641) and less frequently the alanine residues A677 and A687 of the C-terminal EZH2 SET domain. Activating EZH2 mutations lead to an increased formation of trimethylated lysines (H3K27me3), thus disturbing the “histone code.”7-9
Besides mutations, unmutated EZH2 is overexpressed in several types of neoplasms, especially lymphomas. EZH2 overexpression has generally been associated with an adverse prognosis.
Attempts have been made to exploit EZH2 as a therapeutic drug target. Small molecules, designed to inhibit the enzymatic activity of EZH2, showed some promising results.10 These compounds were, however, mainly active against EZH2 with gain-of-function mutations and had little effect in patients with unmutated EZH2. This leads to the hypothesis that there is an additional “noncanonical” oncogenic function of unmutated EZH2, independent of its methyltransferase activity.
In a series of well-designed experiments, Yan and colleagues show that EZH2 also has a transcriptional activator function independent of its histone methyltransferase activity. Starting from the observation that a significant percentage of NKTL patients harbor activating mutations of JAK3, they pursued the hypothesis of a possible functional interaction between EZH2 and JAK3 in NKTL. When investigating coimmunoprecipitates, they observed an association of both proteins. Pharmacologic inhibition of the JAK/STAT5 signaling pathway led to an increased H3K27 trimethylation, suggesting an indirect inhibition of the methyltransferase activity of EZH2 by JAK3. The same effect was observed when JAK3 expression was inhibited by small interfering RNAs, whereas overexpression of JAK3 led to the contrary effect and a decrease in H3K27 trimethylation. Using an anti-phosphotyrosine antibody on immunoprecipitated EZH2, the protein was found to be phosphorylated. This finding was substantiated by the observation that treatment of cells with a JAK inhibitor decreased EZH2 phosphorylation. To identify the precise tyrosine phosphorylation site, the authors bioinformatically identified 3 potential residues (Y23, Y244, and Y523) and generated mutants wherein they substituted tyrosine by alanine. Only the EZH2 Y244A mutant showed significantly reduced cell growth. The Y244A residue was verified as the putative phosphorylation site, using a specific polyclonal antiserum. Treatment of NKTL cells with a JAK3 inhibitor reduced the level of phospho-EZH2 Y244.
Next, the authors investigated the effect of JAK3-mediated phosphorylation of EZH2 on the expression of genes that are downregulated by the histone methyltransferase activity of EZH2. They performed gene expression microarray studies and derived a JAK-STAT activation gene signature and an PRC2-repressed target-gene signature and found a strong positive correlation between the 2 expression signatures.
Using specific EZH2 mutants, the authors identified a set of 93 potential target genes of phosphorylated EZH2 and validated these findings by chromatin immunoprecipitation–quantitative polymerase chain reaction analysis of selected genes. Immunoprecipitation revealed that phosphorylated EZH2 was no longer associated with the PRC2 components SUZ12 and EED (see figure) but instead formed a complex with RNA polymerase II, indicating that the downstream effects of Y244-phosphorylated EZH2 are not dependent on the context of the PRC2.
The application of JAK3 kinase inhibitor PF956980 dramatically decreased NKTL cell growth in cell culture whereas inhibitors of the methyltransferase activity had little or no effect.
In summary, the work of Yan et al reveals interesting new aspects of the molecular biology of EZH2, which was previously known as a methyltransferase and mainly a gene silencer in the context of the PRC2 complex. Yan et al show that EZH2 can also act as transcriptional activator when phosphorylated by JAK3 and that this function is independent and largely exclusive of its methyltransferase activity and the PRC2 complex. Several questions remain to be answered, especially concerning the downstream targets of phosphorylated EZH2. The clinically interesting aspect is of course the perspective that certain patient groups, exhibiting overexpression of (unmutated) EZH2, might benefit from the treatment with JAK inhibitors. There even is the potential of developing specific inhibitors of Y244-phosphorylated EZH2.
Conflict-of-interest disclosure: The author declares no competing financial interests.