To the editor:
Early T-cell precursor acute lymphoblastic leukemia (ETP-ALL) is a recently characterized subtype accounting for up to 15% of childhood T-cell acute lymphoblastic leukemia (T-ALL).1-3 This heterogeneous subgroup shows a distinctive immature immunophenotype characterized by the differential expression of cell surface markers during T-cell development.2,4 ETP-ALL has been associated with a poor prognosis2-6 in both pediatric and adult patients, although this was debated in other studies.7-9
ETP-ALLs are associated with multiple rearrangements affecting gene-coding regions, quite possibly with a bearing on the formation of novel chimeric fusion genes,2,4,10,11 for which the clinical significance remains as yet largely unknown. In order to better understand the factors leading to childhood ETP-ALL relapse, the whole transcriptome of 2 ETP-ALL patients was investigated with a view toward identifying relapse-specific rearrangements.
The 2 patients (TC0002 and TC0022) were diagnosed with ALL at 12 and 16 years of age, respectively, and immunophenotyping revealed antigenic determinants corresponding to ETP-ALL subtype. Both of them were classified as high risk and treated with an l-asparaginase-intensive chemotherapy regimen, according to Dana-Farber Cancer Institute Childhood ALL Consortium protocol for high-risk patients. In both cases, the patients relapsed, after 5 and 2 years respectively, after having obtained an initial remission.
The whole transcriptome analysis revealed a novel recurrent fusion transcript linking the 5′ untranslated region (UTR) of KMT2E to the entire coding sequence of ASNS, indicating a promoter swap (Figure 1A-B). Quantitative reverse transcription polymerase chain reaction (RT-qPCR) was performed at diagnosis, in conjunction with bone marrow samples taken at different time points after induction treatment of TC0002, and these indicated that the fusion was relapse specific (Figure 1C-D). This novel recurrent fusion may have been previously overlooked in other studies as a consequence of the rarity of transcriptome analysis having being performed on relapsed ETP-ALL, as this may indeed be the sole recorded study.
Identification and validation of the KMT2E-ASNS fusion transcripts in 2 relapsed childhood ETP-ALL patients. (A) Fusion transcripts KMT2E-ASNS containing KMT2E exon 1 (5′UTR) linked to the entire coding sequence of ASNS identified in the relapse material of both childhood ETP-ALL patients (TC0002 and TC0022) by RNA sequencing (RNA-seq). The dotted lines indicate observed exon junctions. Filled boxes correspond to coding sequences and small unfilled boxes correspond to UTRs (the first 23 bp of ASNS exon 3 are part of the 5′ UTR). (B) Complementary DNA (cDNA) sequence at exon junctions for the 2 transcript isoforms identified from RNA-seq data. (C) Validation of the fusion transcripts by RT-PCR and Sanger sequencing for both patients. (D) RT-PCR performed on the different time points of 4 samples taken from TC0002s patient on diagnosis, during and after treatment (W5, W16, W108, and W136), and on relapse showing the evolution of the KMT2E-ASNS alteration. KMT2E-ASNS is not detected until relapse (154 weeks after treatment end). D, diagnosis; R, relapse; Tx, treatment; W, week.
Identification and validation of the KMT2E-ASNS fusion transcripts in 2 relapsed childhood ETP-ALL patients. (A) Fusion transcripts KMT2E-ASNS containing KMT2E exon 1 (5′UTR) linked to the entire coding sequence of ASNS identified in the relapse material of both childhood ETP-ALL patients (TC0002 and TC0022) by RNA sequencing (RNA-seq). The dotted lines indicate observed exon junctions. Filled boxes correspond to coding sequences and small unfilled boxes correspond to UTRs (the first 23 bp of ASNS exon 3 are part of the 5′ UTR). (B) Complementary DNA (cDNA) sequence at exon junctions for the 2 transcript isoforms identified from RNA-seq data. (C) Validation of the fusion transcripts by RT-PCR and Sanger sequencing for both patients. (D) RT-PCR performed on the different time points of 4 samples taken from TC0002s patient on diagnosis, during and after treatment (W5, W16, W108, and W136), and on relapse showing the evolution of the KMT2E-ASNS alteration. KMT2E-ASNS is not detected until relapse (154 weeks after treatment end). D, diagnosis; R, relapse; Tx, treatment; W, week.
KMT2E and ASNS are located ∼ 7.1 Mb apart on chromosome 7, in opposite orientations (supplemental Figure 1A, available on the Blood Web site), implying that the observed KMT2E-ASNS transcripts may be the result of a cryptic inversion [inv(7)(q22.3q21.3)]. In spite of the absence of a reciprocal ASNS-KMT2E transcript, we observed a transcript linking the 5′ portion of the MGC72080 pseudogene (located ∼40 kb upstream of ASNS) to exon 2 of KMT2E in the relapse material of both of these patients, which supports the chromosomal inversion hypothesis (supplemental Figure 1B).
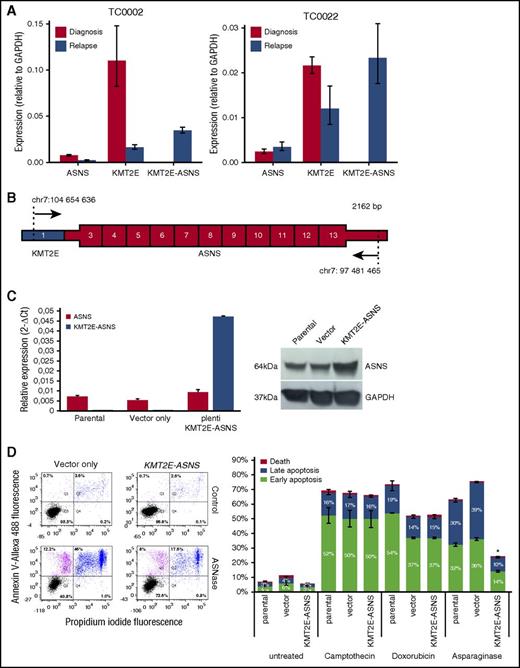
To assess the impact of the promoter swap on ASNS expression, we measured the expression levels of KMT2E, ASNS, and KMT2E-ASNS transcripts in both patients at diagnosis and at relapse for which the procedure has been outlined in the supplemental Methods. The expression levels of KMT2E-ASNS transcripts at relapse are 4.6- and 9.7-fold higher than endogenous ASNS at diagnosis, for TC0002 and TC0022, respectively (Figure 2A).
Relative expression and functional analysis of the KMT2E-ASNS chimera. (A) RT-qPCR analysis of relative expression levels for endogenous KMT2E and ASNS as well as KMT2E-ASNS fusion transcripts in diagnosis and relapse samples of childhood ETP-ALL patients TC0002 and TC0022. Expression levels of KMT2E-ASNS at relapse are 4.6-fold and 9.7-fold higher than endogenous ASNS levels at diagnosis for TC0002 and TC0022, respectively. Expression levels of KMT2E are 6.7-fold and 1.8-fold lower in the relapse samples compared with diagnosis for TC0002 and TC0022, respectively. The vertical bars show 95% confidence intervals according to Student t distribution. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression level was used for reference. (B) Schematic representation of the novel KMT2E-ASNS fusion transcript amplified from TC0002s patient cDNA, subcloned into a pLenti vector and transduced in the MOLT-4 cell line. Exon 1 of KMT2E is shown in blue, and ASNS exons 3 to 13 are shown in red. UTRs are drawn thinner than coding regions. The primers used for amplification are depicted as arrow with the genomic position (hg19) of their first (5′) aligned base. (C) RT-qPCR analysis of relative expression levels for KMT2E-ASNS fusion transcripts in cell transduced MOLT4 cell line. Expression levels of ASNS and KMT2E-ASNS in transduced MOLT4 cell line show a fivefold higher KMT2E-ASNS level compared with wild-type and MOLT4 transduced cell line with vector only. The vertical bars show 95% confidence intervals according to Student t distribution. GAPDH expression level was used for reference. ASNS protein level was shown higher in KMT2E-ASNS transduced cell line compared with wild-type and empty vector’s cell lines (D) Apoptosis assay showing close to 30% reduction in apoptosis in KMT2E-ASNS–positive MOLT-4 cells compared with vector only (pLenti) transduced cells. Apoptosis profile of parental (data not shown) and transduced MOLT-4 cells was determined by flow cytometry using annexin V–Allexa488/ propidium iodide (PI). The numbers in each quadrant indicate the percentage of cells from a total of 10 000 cells. Viable cells appear in the lower left-hand quadrant (PI and annexin V negative), whereas cells in the upper left-hand quadrant and in the upper right-hand quadrant denote early and late apoptotic cells, respectively. The lower right-hand quadrant represents dead cells. Camptothecin and doxorubicin were used as negative control to assess the specificity of l-asparaginase. The apoptosis data presented are representative of 3 independent experiments for each of the 2 biological replicates. *P ≤ .05 according to Mann-Whitney U test for the early and late apoptosis of KMT2E-ASNS cell line compared with empty vector cell line.
Relative expression and functional analysis of the KMT2E-ASNS chimera. (A) RT-qPCR analysis of relative expression levels for endogenous KMT2E and ASNS as well as KMT2E-ASNS fusion transcripts in diagnosis and relapse samples of childhood ETP-ALL patients TC0002 and TC0022. Expression levels of KMT2E-ASNS at relapse are 4.6-fold and 9.7-fold higher than endogenous ASNS levels at diagnosis for TC0002 and TC0022, respectively. Expression levels of KMT2E are 6.7-fold and 1.8-fold lower in the relapse samples compared with diagnosis for TC0002 and TC0022, respectively. The vertical bars show 95% confidence intervals according to Student t distribution. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression level was used for reference. (B) Schematic representation of the novel KMT2E-ASNS fusion transcript amplified from TC0002s patient cDNA, subcloned into a pLenti vector and transduced in the MOLT-4 cell line. Exon 1 of KMT2E is shown in blue, and ASNS exons 3 to 13 are shown in red. UTRs are drawn thinner than coding regions. The primers used for amplification are depicted as arrow with the genomic position (hg19) of their first (5′) aligned base. (C) RT-qPCR analysis of relative expression levels for KMT2E-ASNS fusion transcripts in cell transduced MOLT4 cell line. Expression levels of ASNS and KMT2E-ASNS in transduced MOLT4 cell line show a fivefold higher KMT2E-ASNS level compared with wild-type and MOLT4 transduced cell line with vector only. The vertical bars show 95% confidence intervals according to Student t distribution. GAPDH expression level was used for reference. ASNS protein level was shown higher in KMT2E-ASNS transduced cell line compared with wild-type and empty vector’s cell lines (D) Apoptosis assay showing close to 30% reduction in apoptosis in KMT2E-ASNS–positive MOLT-4 cells compared with vector only (pLenti) transduced cells. Apoptosis profile of parental (data not shown) and transduced MOLT-4 cells was determined by flow cytometry using annexin V–Allexa488/ propidium iodide (PI). The numbers in each quadrant indicate the percentage of cells from a total of 10 000 cells. Viable cells appear in the lower left-hand quadrant (PI and annexin V negative), whereas cells in the upper left-hand quadrant and in the upper right-hand quadrant denote early and late apoptotic cells, respectively. The lower right-hand quadrant represents dead cells. Camptothecin and doxorubicin were used as negative control to assess the specificity of l-asparaginase. The apoptosis data presented are representative of 3 independent experiments for each of the 2 biological replicates. *P ≤ .05 according to Mann-Whitney U test for the early and late apoptosis of KMT2E-ASNS cell line compared with empty vector cell line.
ASNS is involved in the synthesis of asparagine. Increased ASNS levels and gene polymorphisms have been associated with reduced sensitivity to l-asparaginase.12-17 To determine whether the KMT2E-ASNS fusion transcript confers l-asparaginase resistance, we transduced the KMT2E-ASNS fusion into the T-ALL cell line MOLT4 (Figure 2B). KMT2E-ASNS was 5 times more expressed than endogenous ASNS in the transduced cells and both at RNA and protein levels (Figure 2C). l-asparaginase–treated MOLT-4–expressing KMT2E-ASNS showed 30% less apoptosis as when compared with wild-type cells (Figure 2D). No effect on apoptosis was observed following treatment with camptothecin or doxorubicin, suggesting that KMT2E-ASNS has contributed to disease recurrence through l-asparaginase resistance.
In addition to increased ASNS levels in the relapse samples, we observed concomitant decreased expression of KMT2E (Figure 2A), a key regulator of hematopoiesis.18-20 This decrease is likely because of the loss (or inactivation) of 1 KMT2E allele as a result of the chromosomal rearrangement. It has been reported that loss of KMT2E leads to cell cycle arrest and plays a role in myeloid malignancies.18,21 Furthermore, ETP-ALL tumors have been shown to present a high frequency of somatic alterations targeting histone modification genes,4 suggesting that mutations in KMT2E, a histone methyltransferase, might indeed perturb epigenetic regulation and thus contribute to ETP-ALL pathogenesis. Additional studies are required to determine the role, if any, of lower KMT2E expression to resistance and aggressiveness in relapsed childhood ETP-ALL.
In conjunction with the KMT2E-ASNS fusion, our transcriptome analysis also revealed the presence of a cryptic t(10p13;11q21) in both patients that was missed by classical cytogenetics. This alteration is the only other alteration with a probable oncogenic role that we were able to identify from the transcriptome analysis. We confirmed the presence of the resulting known PICALM-MLLT10 fusion (supplemental Figure 2A) in the diagnosis and relapse material of both patients (supplemental Figure 2B) by RT-qPCR. Additional RT-qPCR performed on the bone marrow samples taken at different time points after induction treatment of TC0002 indicated that the fusion was also present at week 5 but was not detected again until relapse (supplemental Figure 2C). The PICALM-MLLT10 fusion, which leads to upregulation of the HOXA gene cluster,22 is observed in 5% to 10% of T-ALL patients.23 It has been characterized by an early stage of maturation arrest and is associated with a poor prognosis24 and treatment resistance or relapse.25 Because the KMT2E-ASNS fusion was only found at relapse in both patients, it is tempting to propose that it was acquired by a PICALM-MLT10–bearing subclone during the initial treatment, leading to its escape from chemotherapy through l-asparaginase resistance. Further investigations could provide valuable insights into the mechanisms underlying their tumor dormancy and late disease recurrence and determine the potential co-occurrence between KMT2E-ASNS and PICALM-MLLT10 in ETP-ALL.
We sought to estimate the frequency of KMT2E-ASNS by performing nested RT-PCR on 12 additional relapsed T-ALL patients for whom we could obtain RNA (4 children and 8 adults) (supplemental Table 1). Among these, 4 adults (P1, P6, P7, P8) could be classified as ETP-ALL. For 7 of these patients, we also had their diagnosis material. The KMT2E-ASNS fusion transcript was not detected in any of these cases. Collectively, and despite the heterogeneous profile of this cohort, these results indicate that KMT2E-ASNS might be restricted to relapsed pediatric ETP-ALL cases. However, additional samples (including newly diagnosed T-ALL cases) and experiments are required to estimate the overall frequency of this fusion and the sensitivity of the assay.
In conclusion, our study provides the first evidence of a somatic rearrangement contributing to acquired chemotherapy resistance and relapse in ETP-ALL through ASNS overexpression. It also highlights the need for further clinical genomic approaches to identify cryptic structural chromosomal abnormalities in childhood leukemia and to gain insight into the biology and pathogenesis of relapse. Additional research would hopefully facilitate clearer decision making and treatment orientation.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors are indebted to the personal involvement of the patients and their parents who fully participated in this study. Patient tissue samples were provided by the Sainte-Justine University Hospital Center Pediatric Cancer biobank. RNA-seq was performed at the Integrated Clinical Genomics Centre in Pediatrics, Sainte-Justine University Hospital Center; Sanger sequencing was performed at the McGill University and Genome Quebec Innovation Centre; and computations were made on the supercomputer Briarée from Université de Montréal, managed by Calcul Québec and Compute Canada. This study was supported by research funds provided by the Ministère de l’Économie, de la Science et de l’Innovation du Québec; the Charles-Bruneau Cancer Center Foundation; Network of Applied Genetic Medicine/Fonds de recherche du Québec-Santé; and the Canadian Institutes of Health Research (D.S.). F.K. is the recipient of a Cole Foundation Fellowship. D.S. holds the François-Karl-Viau Research Chair in Pediatric Oncogenomics. Finally, to the authors thank Haytham Khoury for his involvement with the critical revision of the manuscript.
Contribution: D.S. is the principal investigator and takes primary responsibility for the manuscript: S.L., M. Minden, and C.R. were involved in sample and library preparation for RNA-seq; M.L. performed the bioinformatics analysis; S.L. and F.K. carried out functional assays; F.K., S.L., and M.L. were involved in data integration and analysis; V.S., P.S.-O., and P.B. contributed to data processing; S.C. contributed to serial tracking of gene fusion experiments; F.K., M.L., S.L., J.H., and D.S. wrote the manuscript; S.C., M.K., H.B., and M. Marzouki were involved in critical revision of the manuscript; and all authors approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel Sinnett, Research Center, Sainte-Justine University Hospital, Montréal, QC H3T 1C5, Canada; e-mail: daniel.sinnett@umontreal.ca.
References
Author notes
F.K. and M.L. contributed equally to this study.