Abstract
SCL/TAL1 (stem cell leukemia/T-cell acute lymphoblastic leukemia [T-ALL] 1) is an essential transcription factor in normal and malignant hematopoiesis. It is required for specification of the blood program during development, adult hematopoietic stem cell survival and quiescence, and terminal maturation of select blood lineages. Following ectopic expression, SCL contributes to oncogenesis in T-ALL. Remarkably, SCL’s activities are all mediated through nucleation of a core quaternary protein complex (SCL:E-protein:LMO1/2 [LIM domain only 1 or 2]:LDB1 [LIM domain-binding protein 1]) and dynamic recruitment of conserved combinatorial associations of additional regulators in a lineage- and stage-specific context. The finely tuned control of SCL’s regulatory functions (lineage priming, activation, and repression of gene expression programs) provides insight into fundamental developmental and transcriptional mechanisms, and highlights mechanistic parallels between normal and oncogenic processes. Importantly, recent discoveries are paving the way to the development of innovative therapeutic opportunities in SCL+ T-ALL.
Introduction
Lineage specification, commitment, and differentiation are essential biological processes underlying all cellular pathways. They involve, among other regulatory mechanisms, progressive acquisition of tissue-specific programs of gene expression. In hematopoiesis, this directs specification of mesoderm into blood-fated cells, production of hematopoietic stem cells (HSCs), and differentiation into highly specialized blood cells. Emphasizing the intricate control of these processes, their deregulation often leads to aberrant cell proliferation/differentiation and cancer.
Transcription factors (TFs) are essential effectors at the end of a cascade of extracellular and intracellular regulatory mechanisms that establish gene expression networks conferring and progressively sealing cell fates. The basic helix-loop-helix (bHLH) protein stem cell leukemia (SCL)/T-cell acute lymphoblastic leukemia (T-ALL) 1 (TAL1) (hereafter referred to as SCL) is a pivotal hematopoietic transcriptional regulator. This protein not only lies at the apex of the hierarchy of TFs involved in hematopoietic specification, but is also required in adult HSCs and for terminal maturation of select blood lineages. When ectopically expressed, it is involved in the physiopathology of T-ALL. Mechanistically, SCL engages with a large array of protein partners to establish activating and repressive transcriptional activities in a lineage- and stage-specific manner. At the heart of these regulatory complexes are conserved combinatorial protein associations and a subtle interplay of DNA-binding activities. Therefore, studying SCL provides an excellent window into the versatility of TFs and their modular responses to distinct cellular environments, both in normal and malignant milieus.
Here, we review the function and mechanisms of action of SCL in mesoderm to HSCs, blood lineages, and leukemia, contrasting its activities in specification, maturation, and oncogenic processes. We discuss how these findings are shaping the development of targeted drug therapies.
SCL confers hematopoietic fate to mesodermal/endothelial precursor cells
The SCL gene was cloned by virtue of its involvement in chromosomal translocation t(1;14)(p33;q11) from a cell line derived from a patient presenting with T-ALL.1 Because of its capacity to differentiate into lymphoid and myeloid lineages, the leukemia cell line was referred to as a “stem cell” line and the newly cloned gene was termed “stem cell leukemia.” Since then, studies have progressively unveiled SCL’s critical mechanistic role in normal and malignant hemopoiesis, confirming initial hypotheses about parallel functions in HSC development and leukemic transformation.
In vivo ablation of SCL activity in murine models provided the first evidence that SCL functions at early stages of blood development. Scl−/− embryos died at day embryonic day 9.5 (E9.5) from absence of yolk sac (YS) primitive erythropoiesis and myelopoiesis (wave 1, see Box 1).2,3 Moreover, all adult definitive hematopoietic lineages (wave 3) were absent in Scl−/− mouse chimeras.4,5 This complete block in hematopoiesis suggested a function in either the first differentiation steps from blood stem/progenitor cells or the specification of mesodermal cells toward a blood fate.
Box 1.
Blood development: the 3 waves
During vertebrate embryogenesis, blood development occurs in 3 successive waves (Figure 1).6,8 The first 2 waves (1 and 2) take place in the extraembryonic YS and give rise to transient blood populations; the third wave (3) develops in the intraembryonic aorta-gonado-mesonephros (AGM) region and gives rise to hematopoietic stem/progenitor cells (HSPCs), providing the organism with lifelong blood production. While wave 1 produces primitive red blood cells (expressing embryonic globins), megakaryocytes, and macrophages, waves 2 and 3 give rise to definitive multipotent cells: erythromyeloid progenitors (EMPs) in the YS blood islands, and HSPCs in the dorsal aorta (DA), respectively. Waves 2 and 3 produce a highly specialized endothelium, referred to as hemogenic endothelium (HE), from which these hematopoietic progenitors bud in a process known as the endothelial-to-hematopoietic transition (EHT).9-12 This is consistent with the hypothesis, first framed 100 years ago, of an endothelial origin for blood cells.13 EMPs and HSPCs subsequently migrate to and differentiate in the fetal liver to produce adult-type blood cells.
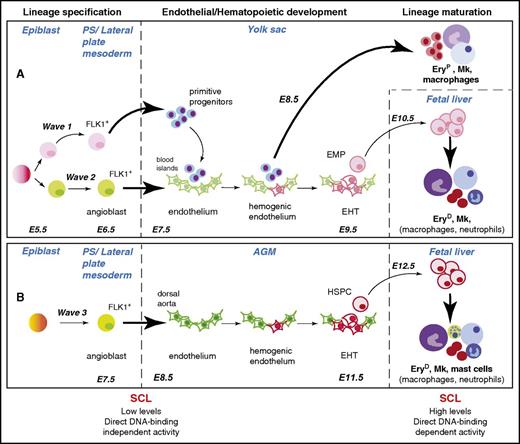
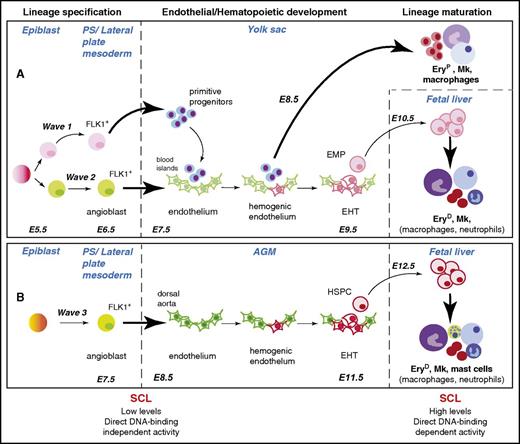
SCL is required for development of the 3 hematopoietic waves at specification and maturation stages. Specification, development, and maturation stages of hematopoietic waves 1, 2, and 3 during mouse development are depicted.6 A common origin for waves 1 and 2 in the early epiblast is shown, but this is still the subject of debate (see “SCL confers hematopoietic fate to mesodermal/endothelial precursor cells” and Box 2). The (A) YS and (B) aorta-gonad-mesonephros (AGM) waves are shown as independently specified, as established in Xenopus embryos through elegant lineage-tracing studies.7 However, the origin of the angioblasts giving rise to wave 3 is not yet established in higher vertebrates. The SCL-dependent cellular transitions are represented by bold arrows. The hematopoietic lineages described as dependent on SCL activity for terminal maturation in the fetal liver are in bold font. The main features of SCL’s activities in specification and maturation stages are summarized below the diagram. E5.5-E12.5, embryonic days E5.5-E12.5; EHT, endothelial-to-hematopoietic transition; EryD, definitive erythroid cells; EryP, primitive erythroid cells; Mk, megakaryocyte; PS, primitive streak.
SCL is required for development of the 3 hematopoietic waves at specification and maturation stages. Specification, development, and maturation stages of hematopoietic waves 1, 2, and 3 during mouse development are depicted.6 A common origin for waves 1 and 2 in the early epiblast is shown, but this is still the subject of debate (see “SCL confers hematopoietic fate to mesodermal/endothelial precursor cells” and Box 2). The (A) YS and (B) aorta-gonad-mesonephros (AGM) waves are shown as independently specified, as established in Xenopus embryos through elegant lineage-tracing studies.7 However, the origin of the angioblasts giving rise to wave 3 is not yet established in higher vertebrates. The SCL-dependent cellular transitions are represented by bold arrows. The hematopoietic lineages described as dependent on SCL activity for terminal maturation in the fetal liver are in bold font. The main features of SCL’s activities in specification and maturation stages are summarized below the diagram. E5.5-E12.5, embryonic days E5.5-E12.5; EHT, endothelial-to-hematopoietic transition; EryD, definitive erythroid cells; EryP, primitive erythroid cells; Mk, megakaryocyte; PS, primitive streak.
Subsequent studies in series of experimental models pointed to a role for SCL in endothelial and blood development rather than from already established blood progenitors. Immunolabeling revealed SCL expression in (1) dispersed mesodermal endothelial cell precursors (angioblasts) expressing fetal liver kinase-1 (FLK1; receptor for vascular endothelial growth factor A [VEFGA]) in E6.5 to E7.5 mouse embryos, (2) angioblasts in the splanchnic mesoderm of avian embryos, and (3) FLK1− hematopoietic cells in mouse YS blood islands.14,15 Enforced SCL expression in zebrafish and Xenopus embryos was sufficient to induce mesoderm to hematopoietic and endothelial fates.16,17 Finally, differentiation of Scl-null cells in murine embryonic stem (ES) cell models, which recapitulate waves 1 and 2 of YS hematopoiesis,6 established that SCL was required for generation of both blood and endothelial components of colonies derived from FLK1+ mesodermal cells (also called blast colony-forming cells).18,19 Taken together, this suggested a role for SCL at the onset of blood/endothelial development.
Conditional deletion of Scl through Tie2-Cre recombinase-mediated panendothelial excision refined these observations.20 In this model, hematopoietic specification was not affected, but maturation of primitive and definitive erythroid cells as well as megakaryocytes was defective, leading to lethality at days E13.5/14.5. This study informed on 3 key aspects of SCL’s functions: (1) SCL is required before Tie2-Cre becomes functional, (2) SCL activity in blood fate specification can be uncoupled from a later role in blood cell maturation, and (3) after specification of the blood lineage, SCL is not required for HSPC production (Figure 1).
SCL is also necessary for development of the vascular network after angioblast formation. Ablation of Scl expression disrupted extraembryonic angiogenic remodeling in mouse embryos.21,22 In zebrafish and Xenopus, Scl loss led to disorganization of major blood vessels, including the DA.8,23,24
In summary, during development, SCL is essential for (1) specification of the 3 hematopoietic waves, (2) maturation of select blood lineages, and (3) remodeling of the vascular network.
In which cell types does SCL exert its function? Regarding extraembryonic hematopoiesis, the prevailing hypothesis is that SCL acts in the precursor of YS blood and endothelial populations giving rise to waves 1 and 2, the hemangioblast, located in the primitive streak (PS). However, recent lineage-tracing studies propose independent origins for blood and endothelial lineages, in the epiblast, before gastrulation and formation of the PS (see Box 2).11 This supports earlier fate-mapping studies describing ingression of erythroid-fated cells prior to endothelial-fated cells into the PS of gastrulating embryos.25 Of note, lineage restriction of cardiovascular progenitors is also believed to occur before gastrulation.26 Together, this suggests that mesoderm patterning and lineage specification decisions might take place pre-PS formation. Importantly, this model does not exclude a common origin for progenitors of waves 1 and 2, but in early epiblast stages, before restriction of potentiality and regional segregation as cells ingress into the PS.11,25 Further lineage-tracing studies are required to identify this cell and refine our understanding of blood fate determination.
Box 2.
The hemangioblast: toward a new definition?
The hemangioblast was first defined a century ago as a cell giving rise to both blood and endothelial lineages in the YS of chick embryos.27,28 This cell was later identified in the PS of mouse embryos and described as a bi- (blood/endothelial) or tri- (blood/endothelial/vascular smooth muscle) potential clonal progenitor both in vitro and in vivo.29,30 However, a recent retrospective clonal lineage-tracing study disputes this model and reports independent blood and endothelial progenitors in the epiblast, before formation of the PS and migration of cells to the YS.11 This finding, although not excluding the existence of a common blood and endothelial progenitor before separation of the 2 lineages in the epiblast, questions the existence of the hemangioblast as originally defined.
Therefore, we propose that SCL acts on the progenitors of the YS primitive blood cells (wave 1) and blood-fated angioblasts (wave 2) after their ingression into the PS and before their migration into the YS where they form the blood islands (Figure 1). Similarly, in this model, SCL is required to instruct the angioblasts at the origin of wave 3 (as demonstrated in Xenopus embryos, see “SCL lies at the top of the hematopoietic transcriptional hierarchy”). Full examination of the function and spatiotemporal expression of SCL in early stages of gastrulation is required to confirm this model.
Vascular endothelium, cardiomyocytes, and endocardium: an intricate relationship
Manipulation of SCL expression during development has highlighted complex relationships between endothelial cells and the cardiovascular lineage.
Blood-fated vascular endothelium and cardiomyocytes
In zebrafish embryos, forced expression of Scl messenger RNA (mRNA) expanded blood and endothelial tissues at the expense of somitic and myocardial tissues.16,24,31 Conversely, downregulation of Scl led to ectopic cardiomyocyte production and unveiled fibroblast growth factor–mediated repression of a latent cardiac potential in blood and vessel progenitors.32,33 In agreement with this, absence of SCL revealed cardiac potential in FLK1+ blood-fated endothelial cells of murine YS.34 Corroborating these phenotypes, SCL not only activates vascular- and blood-specific transcriptional programs in FLK1+ cells, but also represses expression of cardiac-specific regulators and structural proteins to prevent ectopic cardiac activity (Org et al35 and H.C., M.S.K., and C.P., SCL establishes a repressive environment in blood-fated cells, unpublished data).
Endocardium and myocardium
Although endocardial precursors are specified in Scl mutant embryos, endocardial cell migration and maturation are impaired, leading to defective intercellular junctions and heart tube formation.36,37 Interestingly, absence of SCL in the endocardium also induces ectopic myocardial differentiation.34,37 Therefore, as for vascular endothelium, SCL is required for the biology and identity of the endocardium.
Conclusions
Altogether, these studies open a window into intricate lineage relationships in early embryogenesis and developmental plasticity. Expression of SCL in vascular endothelium and endocardium suggests a shared origin from a common endothelial progenitor. Separately, the requirement for SCL to restrict a cardiomyocyte fate in blood-fated vascular endothelial cells and endocardium suggests a developmental relationship between myocardium and endothelial cells. Whether the cardiovascular and blood/endothelial systems share a common origin is, however, still under debate.26,38-40 Understanding the detailed mechanistic basis to these processes is likely to provide principles to a recurring question in developmental biology: at what stage do lineage-fated cells arise and what is the extent of multipotentiality of common progenitors?
SCL lies at the top of the hematopoietic transcriptional hierarchy
Studies of other TFs required in early stages of mesoderm/blood development have helped position SCL in the hierarchy of regulatory events leading to endothelial and blood specification. An important regulator of vascular and hematopoietic development is the ETS family TF, ETS variant 2 (ETV2). Etv2−/− mouse embryos die at E9.5/10.5 from the complete absence of vasculature and YS hematopoiesis,41 inferring a role in mesoderm specification toward blood/endothelial lineages. Indeed, in response to VEGFA signaling, ETV2 is transiently required for progression of FLK1+PGFRα+ early mesoderm to FLK1+PDGFRα− lateral plate mesoderm, the population containing the angioblasts.42-45 ETV2 is believed to control segregation of a heterogeneous population of angioblasts into distinct lineages through differential activation of genetic programs: ETV2’s target gene miR-130a directs angioblasts toward endothelial cells, but not blood-fated endothelial cells,46 whereas Scl and the transcriptional regulators Gata2 and Fli1 (ETS family), both also involved in endothelium/blood development, are critical for induction of blood-fated cells.42,44,47,48 As SCL, GATA-binding protein 2 (GATA2), and friend leukemia integration 1 (FLI1) participate in an interconnected regulatory loop,49 they are able to sustain a genetic program after the initiating events (VEGFA-ETV2) have ceased. This mechanism, known to specify and maintain cell identity,50 confers hematopoietic fate to angioblasts at the origin of the blood lineage.
We are now starting to appreciate aspects of SCL function in angioblasts giving rise to the adult HSPC lineage, from studies in Xenopus embryos. Here, ETV2, ETV6 (another ETS protein), and VEGFA signaling initiate expression of Scl and additional endothelial and blood markers.51 Upon acquisition of an endothelial identity, the angioblasts migrate from the dorsal lateral plate (DLP) mesoderm toward the embryo midline where they form the DA. Knockdown of Scl negatively affects expression of blood/endothelial genes in the DLP. This does not prevent endothelial cell migration, but disrupts DA formation and hematopoietic development.8,51 As Scl mRNA expression is downregulated in endothelial cells migrating toward the embryo midline,7 SCL is likely to instruct endothelial cell progenitors in the DLP before their migration. The mechanisms of how SCL directs these processes are not clear but may involve the establishment of appropriate epigenetic/transcriptional landscapes. Indeed, recent genome-wide chromatin immunoprecipitation (ChIP) analyses of ES cell-derived cellular intermediates have shown SCL binding to genomic targets in GFP-Brachyury+/FLK1+ cells, prior to endothelial development and appearance of DNase I–hypersensitive sites.52 This suggests that SCL may transcriptionally prime key target loci in DLP cells for activation at a later time: these become expressed at high levels at the time of DA formation and HE production, possibly through remodeling of TF-binding patterns by factors such as Runt-related TF 1 (RUNX1), the key regulator of EHT processes (this issue of Blood and Lichtinger et al53 ).
Therefore, SCL is one of the early-acting hematopoietic TFs likely to prime cis-acting elements in gene loci involved in hemopoietic and endothelial development.
SCL in adult hematopoiesis
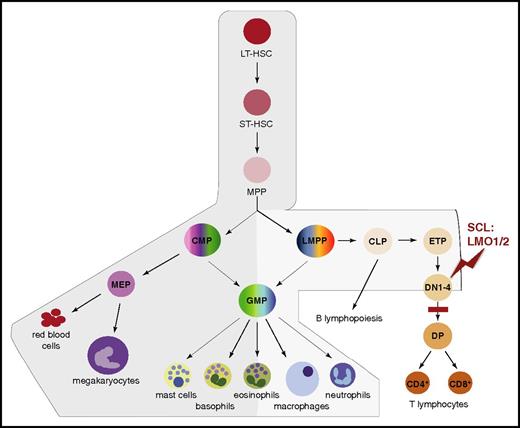
Although broadly expressed in adult blood cells, Scl mRNA levels mark distinct branches of the hematopoietic tree. Scl is expressed in the HSC compartment, myeloid progenitors, and mature myeloid cells (red cells, megakaryocytes, mast cells, and basophils; at lower levels in eosinophils, macrophages, and neutrophils) and at low levels in lymphoid progenitors. In contrast, Scl is absent in differentiating T and B lymphocytes (Figure 2).56-58 As described in the following paragraphs and in “SCL in leukemogenesis,” this pattern of expression reflects SCL’s requirements in HSCs and myeloid differentiation, and its nonpermissive role in the lymphoid compartment.
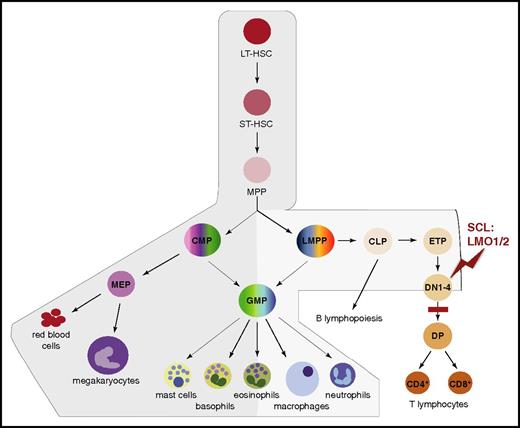
Scl mRNA domains of expression mark distinct branches of the hematopoietic tree. Schematic representation of the successive stages leading to hematopoietic differentiation from LT-HSCs to mature blood lineages.54 The branches exhibiting expression of Scl mRNA are shaded in gray, with darker gray representing higher expression levels. During T-cell differentiation, SCL is downregulated at the DN3 stage. The stage of T-cell development first sensitive to SCL:LMO1/2 ectopic expression (red zigzag) and the subsequent block in differentiation (red bar) in T-ALL mouse models are shown. In human patients, maturation arrest occurs at the late cortical DP CD3high stage (not shown).55 CLP, common lymphoid progenitor; CMP, common myeloid progenitor; DN1-4, CD4−CD8− double-negative populations, stages 1 to 4; DP, CD4+CD8+ double-positive population; ETP, early thymic progenitor; GMP, granulocyte/macrophage progenitor; LMPP, lymphoid/myeloid-primed progenitor; LT-HSC, long-term HSC; MEP, megakaryocytic/erythroid progenitor; MPP, multipotent progenitor; ST-HSC, short-term HSC.
Scl mRNA domains of expression mark distinct branches of the hematopoietic tree. Schematic representation of the successive stages leading to hematopoietic differentiation from LT-HSCs to mature blood lineages.54 The branches exhibiting expression of Scl mRNA are shaded in gray, with darker gray representing higher expression levels. During T-cell differentiation, SCL is downregulated at the DN3 stage. The stage of T-cell development first sensitive to SCL:LMO1/2 ectopic expression (red zigzag) and the subsequent block in differentiation (red bar) in T-ALL mouse models are shown. In human patients, maturation arrest occurs at the late cortical DP CD3high stage (not shown).55 CLP, common lymphoid progenitor; CMP, common myeloid progenitor; DN1-4, CD4−CD8− double-negative populations, stages 1 to 4; DP, CD4+CD8+ double-positive population; ETP, early thymic progenitor; GMP, granulocyte/macrophage progenitor; LMPP, lymphoid/myeloid-primed progenitor; LT-HSC, long-term HSC; MEP, megakaryocytic/erythroid progenitor; MPP, multipotent progenitor; ST-HSC, short-term HSC.
SCL and HSCs: survival and quiescence
SCL’s functions in adult long-term repopulating HSCs have been complex to decipher,59-61 as these roles are shared with another bHLH protein, lymphoblastic leukemia 1 (LYL1). Although LYL1 does not compensate for SCL in specification processes,62,63 Scl/Lyl1 conditional double knockout mouse models revealed redundant, antiapoptotic, and dose-dependent functions in HSC survival.64 Using transplantation assays in proliferative stress conditions, Lacombe et al were then able to demonstrate a gene dosage effect in Scl+/− mouse bone marrow HSCs. SCL negatively regulates the G0-G1 transition in adult Kit+/Lin−/Sca1+ (KLS)/CD150+/CD48+ HSCs, in part by controlling expression of the genes encoding TF inhibitor of DNA binding 1 (ID1) and cyclin-dependent kinase (CDK) inhibitor P21/CDK inhibitor 1A (CDKN1A).65 HSC quiescence and long-term activity are further maintained through a positive feedback loop involving the cytokine receptor c-KIT.66 Interestingly, SCL exhibits opposite functions in human cord blood HSCs where it is reported to positively control proliferation through the mammalian target of rapamycin pathway.67 In conclusion, these studies have highlighted an important facet of SCL in survival and self-renewal mechanisms to preserve adult HSC integrity. They have also revealed SCL's ontogeny-specific activities, underlining some of the developmentally-related distinctive features of HSCs: proliferative (embryonic HSCs) vs quiescent (adult HSCs).
SCL and adult endothelium: a role in vascular repair
Reminiscent of its functions in vascular remodeling during development, SCL controls postnatal angiogenesis. Although low in quiescent endothelial cells, its expression is upregulated during morphogenesis events.68 Mechanistically, SCL regulates the development of human adult endothelial progenitors through activation of genes involved in cell adhesion and migration; this property has been exploited to confer vascular repair functions to endothelial cells in transplantation settings, opening the way to therapeutic applications.69
SCL and lineage maturation: balance between proliferation and differentiation
Gain- and loss-of-function studies have highlighted SCL’s roles in lineage progenitors and their differentiation into erythroid cells, megakaryocytes, mast cells, and monocytes/macrophages.20,70-73 In contrast to adult HSCs, SCL activates cell cycle progression in progenitors. Through tight control of p21 and p16/Ink4a expression, SCL regulates the onset of red cell differentiation, polyploidization and terminal maturation of megakaryocytes, as well as proliferation of monocyte progenitors.72-74 SCL also promotes cell survival through antiapoptotic activities. In physiological conditions requiring control of red cell production, caspase-mediated cleavage of SCL leads to reduced expression of TF GATA1 and survival factor BCL-XL, thereby triggering apoptosis.75
Genome-wide interrogation of SCL’s direct target genes provided further insight into its activities in lineage differentiation. ChIP sequencing combined with gene expression analyses from primary erythroblasts revealed that SCL controls both general processes (transcription, cell cycle, proliferation) and erythroid-specific pathways (redox processes, heme biosynthesis, cytoskeleton organization).76 Comparative studies of SCL ChIP-sequencing data from multipotent blood progenitors, proliferative lineage-specific precursors, and terminally differentiating erythroid and megakaryocytic cells reported dramatic changes in SCL occupancy through the progressive stages of commitment and differentiation.77 In progenitor cells, SCL target genes were enriched in processes associated with proliferation and apoptosis. Megakaryocytic genes were bound by SCL in progenitor cells, suggesting priming of genetic loci associated with low-level expression, as in mesodermal precursors. In contrast, erythroid-specific genes were typically bound in the erythroid lineage only, but the binding pattern of SCL varied as the cells matured, often reflecting changes in levels of target gene expression.77 These dynamic changes suggest a central role in lineage commitment and terminal maturation. What drives these processes is unclear, but they likely stem from the adaptable nature of SCL-containing transcriptional complexes.
SCL and multiprotein regulatory complexes: an elaborate chemistry
To fully appreciate SCL’s mechanisms of action, it is essential to consider SCL-containing combinatorial multiprotein complexes and their recruitment to genomic targets.
SCL’s DNA-binding activity is not the primary determinant of genome occupancy
SCL binds the consensus Ebox DNA motif, CANNTG, as a heterodimer with ubiquitously expressed bHLH E-proteins (E12/E47, HEB, E2-2).78 Non-DNA-binding proteins (bridging LMO [Lin11, Isl-1, and Mec-3 (LIM)-only] and LDB [LIM domain-binding] proteins) are then recruited to form the SCL core complex: SCL:E-protein:LMO1/2:LDB1. This quaternary module, described in all SCL-expressing cells, recruits additional DNA-bound TFs and cofactors. Among the best-described partners are the GATA proteins: GATA2, involved in HSPC specification/survival and the early stages of lineage differentiation, and GATA1, required in lineage maturation.47,79,80 In vitro enrichment of DNA-binding sites initially demonstrated the assembly of the pentameric complex (SCL:E47:LMO2:LDB1:GATA1) on a bipartite Ebox-GATA DNA motif.81 Interestingly, analyses of the sequences underlying SCL ChIP-sequencing peaks invariably describe GATA motifs as the most frequent SCL-bound sequences.35,76,77,82 Therefore, throughout development, GATA proteins appear as a major determinant of SCL genomic occupancy. Together with the identification of 1/2 Ebox-GATA motifs in sequences underlying SCL ChIP peaks (CTG(N9-10)A/WGATAA, the 1/2 Ebox CTG being bound by E47),76,83,84 this put into perspective the importance of Ebox motifs in recruiting SCL-containing complexes.
SCL bridges transcriptional activities to other DNA-bound regulators
The x-ray structure of the SCL quaternary complex (SCL:E47:LMO2:LDB1) bound to an Ebox revealed how binding of LMO2 strengthens the interactions between SCL and E47 through creation of new hydrogen bonds.83,85 This, in turn, induces a rotation of E47 that weakens the affinity of the heterodimer for DNA. Through its adaptor function, LMO2 links the heterodimer to other DNA-bound proteins, such as the GATA proteins,83 leading to recruitment of SCL:E-protein:LMO2 complexes to GATA1/2-bound genomic loci. Interaction with additional DNA-bound proteins therefore provides DNA-binding specificity, more so than Ebox or 1/2 Ebox motifs, the latter merely acting as an anchoring point for the heterodimer when associated to GATA sites. Altogether, this explains, in part, why some of SCL’s functions, especially in blood specification, are independent of SCL’s own ability to bind DNA.63,76,86 As cofactors such as p300/CBP and ETO2 (and possibly mSIN3A and pCAF) interact directly with E-proteins,83,87 SCL’s main role may be to act as a linker connecting ubiquitously expressed E-proteins and associated transcriptional activities to tissue-specific DNA-binding proteins through interaction with LMO2 (Figure 3). This is supported by structure/function studies showing that only the helix-loop-helix domain of SCL (mediating both heterodimerization and LMO2 binding) is indispensable for functional rescue of blood specification in an Scl−/− background.63 Finally, the SCL complex is involved in chromatin architecture through DNA looping. In erythropoiesis, looping occurs upon dimerization of LDB1 to juxtapose enhancers and promoters. This triggers recruitment of cofactors and RNA polymerase II at promoters, leading to gene activation.88,89
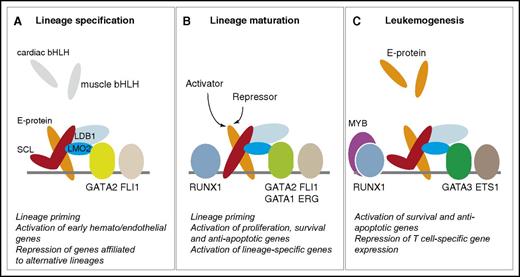
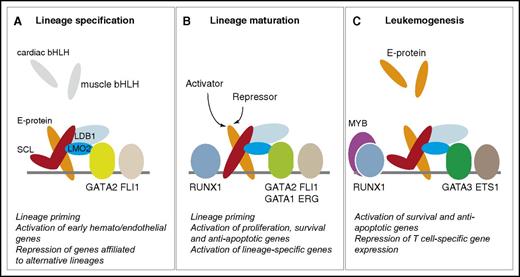
SCL-containing multiprotein complexes in hematopoiesis and leukemogenesis. The quaternary complex (SCL:E-protein:LMO2:LDB1) interacts with members of the GATA, ETS, and RUNX families throughout hematopoietic development and in leukemogenesis. The “kinked” SCL molecule represents a DNA-binding independent form of SCL. (A-C) The main functions of SCL in each compartment are summarized under each diagram, as detailed throughout the text.
SCL-containing multiprotein complexes in hematopoiesis and leukemogenesis. The quaternary complex (SCL:E-protein:LMO2:LDB1) interacts with members of the GATA, ETS, and RUNX families throughout hematopoietic development and in leukemogenesis. The “kinked” SCL molecule represents a DNA-binding independent form of SCL. (A-C) The main functions of SCL in each compartment are summarized under each diagram, as detailed throughout the text.
bHLH, GATA, ETS, and RUNX proteins: the winning combination
Following the observation that SCL/GATA2/FLI1 coordinately control hematopoietic development, parallel analyses of TF ChIP-sequencing assays identified a similar combination of regulators in adult hematopoiesis. A “heptad” of TFs, namely SCL, LYL1, GATA2, FLI1 and ERG (ETS proteins), RUNX1, and LMO2, function cooperatively in HSPCs and cell intermediates as they differentiate into erythroblasts and megakaryocytes.90,91 Although composition of the protein complexes binding to regulatory elements at any one time still remains to be identified, pairwise combination and motif search analyses demonstrated the combinatorial nature of TF interactions. This has unveiled previously uncharacterized combinations, such as SCL/RUNX1, and a strong GATA/ETS correlation in most genomic regions bound by the heptad.91,92 Comparative analyses of the megakaryocytic and erythroid lineages suggested overlapping and divergent roles for these 7 TFs, thus predicting a role in lineage branching. They highlighted distinct binding of SCL, distinct combinations of TF-binding motifs, and distinct patterns of GATA1 and GATA2 binding in the 2 lineage branches. The switch in GATA1/GATA2 binding is likely to play an important role in the shifts in SCL genomic occupancy and lineage decisions,77,90 possibly associated with differential use of SCL short and long isoforms.93 Altogether, these data reinforce the critical requirement for the bHLH/GATA/ETS protein triad throughout hematopoietic development that also involves RUNX1 in HSPCs and megakaryocytes.
The pivotal role of SCL-containing complexes in specification and differentiation processes was highlighted in recent reprogramming experiments. Forced expression of SCL with various combinations of GATA1/2, ERG, RUNX1, and LMO2 induces differentiation of fibroblasts and pluripotent stem cells into multipotent,52,94 erythroid (with cMYC),95 or erythromegakaryocytic96 progenitors through an HE intermediate (reviewed in Hoang et al97 ). Direct fate conversion reflects the capacity of these regulators to overcome epigenetic barriers and robustly establish lineage-specific transcriptional programs, suggesting that some of them may act as pioneering factors or interact with such factors.98
Repressive vs activating functions
In addition to activation of gene expression, repression mechanisms play an increasingly recognized role in lineage determination. This reflects the fact that (1) unilineage-fated cells derive from multilineage-primed progenitors and, therefore, need to establish repressive mechanisms to adopt a unique cell fate and (2) genes required for terminal maturation are often primed in early stages of lineage commitment but their expression needs to be restrained to prevent premature high-level expression and precocious differentiation. During development, SCL prevents expression of cardiac-affiliated genes through active repressive mechanisms (H.C., M.S.K., and C.P., SCL establishes a repressive environment in blood-fated cells, unpublished data) and by occupation of cardiac enhancers hampering activation by cardiac-specific TFs.35 In red cells, SCL interacts with an extended network of corepressors, comprising ETO2/GFI1B/NCOR1/mSIN3A, to repress primed genes.74,99,100 Subsequent gene activation necessitates interaction with coactivators, such as p300/CBP and pCAF. The shift from repressive to activating complexes has been documented for ETO2 and P300 where competitive binding to the AD1 domain of E-proteins regulates complex formation.87 Moreover, the nature of other protein partners, such as GATA proteins, influences multiprotein complex transcriptional activity. In erythropoiesis, SCL complexes exhibit repressive properties in progenitors through association with GATA2, whereas interaction with GATA1 activates genes required for terminal maturation.80,90,101
Conclusions
Altogether, these studies invite further identification of the mechanisms orchestrating dynamic recruitment of regulatory complexes to their genomic targets as hematopoietic differentiation proceeds. In particular, the relationship between SCL and chromatin structure77 merits investigation at a mechanistic level. Changes in the epigenetic landscape, upon expression/repression of pivotal regulators, could generate an environment propitious to de novo enhancer formation or eradication of existing enhancers, leading to TF-binding pattern remodeling and alteration of gene expression programs.
Specification vs maturation
A parallel can be drawn between SCL’s functions and (1) its DNA-binding activities, (2) expression levels, and (3) protein isoforms. Studies using Scl hypomorphic zebrafish embryos and mouse embryos expressing a DNA-binding mutant form of SCL have shown that low levels of SCL and DNA-binding independent activities were sufficient for lineage specification. Conversely, higher levels and direct DNA-binding activities were required for lineage maturation.86,102 In zebrafish, the SCL isoform required in HSPC specification is subject to rapid degradation resulting in low protein levels.103 This suggests that low levels and DNA-binding independent activities of SCL are sufficient for lineage priming and gene repression in specification processes, and may be a prerequisite to prevent precocious hematopoietic differentiation in blood-fated angioblasts. In contrast, higher levels of SCL protein together with direct DNA-binding activities are required for robust gene expression of primed genes in differentiation processes (Figure 1).
SCL in leukemogenesis
Aberrant expression of transcriptional regulators often triggers oncogenic processes. In normal conditions, SCL expression is downregulated during T-cell differentiation (Figure 2).104 However, SCL expression can be activated in T cells through chromosomal translocations (interstitial deletions in the SIL-SCL locus) and mono/biallelic transcriptional mechanisms.105 This ectopic thymic expression is seen in 60% of childhood and adult T-ALL cases, often associated with poor prognosis.55,106 Expression of SCL is, however, insufficient to induce overt leukemia. Collaboration with additional oncogenic events is required for full leukemic transformation with short latency periods. In ∼45% of SCL+ T-ALL cases, the cooperating genetic event is ectopic expression of LMO1 or LMO2 through chromosomal rearrangement.55 Murine transgenic models have confirmed this cooperation and shown that SCL and LMO1/2 coexpression confers aberrant self-renewal to CD4−CD8− double-negative (DN3) preleukemic thymocytes (Figure 2).107-109 This self-renewal capacity is enhanced by activation of a major contributor to T-ALL, the NOTCH pathway.110,111 This leads to acquisition of additional mutations, differentiation arrest, and full-blown leukemia.97,107
Parallels with normal hematopoiesis
Remarkably, the oncogenic SCL protein complexes are replicas of those observed in normal hematopoiesis. This occurs as genetic mutations activating Scl result in wild-type SCL protein expression. Moreover, members of the protein families normally interacting with SCL are co-expressed in T cells, either endogenously (RUNX1/GATA3) or ectopically (LMO1/2). Recent genome-wide and structural approaches have refined our understanding of SCL’s mechanisms of action in T-ALL and provided further compelling evidence for parallel functions in hematopoiesis and leukemogenesis.
Direct DNA-binding independent mechanisms, sequestration, and relocation.
In normal thymocytes, E-protein homodimers direct progression of T-cell differentiation through activation of tissue-specific genes.112 When ectopically expressed, SCL sequesters E-proteins in heterodimers.104,113 As structural studies have shown, recruitment of LMO1/2 stabilizes heterodimerization, reinforcing a shift in equilibrium from E-protein homodimers to more stable heterodimers.83 Because of weaker interactions at the heterodimer:DNA interface,83 SCL:E-protein:LMO1/2 complexes are directed to new sets of genomic targets through interaction with additional DNA-bound regulators, such as GATA3, ETS proteins, or RUNX1. This results in repression of proapoptotic and T-cell differentiation transcriptional programs and activation of self-renewal and antiapoptotic genes.82,104,114-117 This sequestration/relocation model is particularly relevant as, as in hematopoietic specification,86 SCL’s mechanisms of action in T-ALL do not require direct DNA-binding activities.117 By analogy, in specification processes, SCL may sequester E-proteins in blood-fated cells away from cardiac or paraxial bHLH proteins to favor a hematopoietic gene expression program and prevent promiscuous development of alternative lineages (Figure 3).
Autoregulatory interconnected loops: SCL, GATA3, RUNX1, ETS1, and MYB.
In T-ALL, SCL, GATA3, and RUNX1 autoregulate each other and positively control expression of key target genes, such as MYB which, in turn, contributes to maintaining the oncogenic transcriptional program.116 The discovery of mutations creating a de novo MYB-binding site in the SCL locus and triggering formation of a broad enhancer (termed superenhancer) not only provided a genetic mechanism for SCL monoallelic expression, but also helped refine the composition of SCL regulatory complexes.118 Mechanistically, MYB binding drives SCL autoregulation through recruitment of CBP, broad H3K27 acetylation, chromatin opening, and nucleation of SCL-containing multiprotein complexes involving RUNX1, GATA3, and ETS1. MYB/CBP association, together with SCL, GATA3, and RUNX1, positively regulates transcription of each component of the complex, thus placing MYB in the regulatory kernel. It would be interesting to determine whether MYB is also part of the recursive loops documented in normal hematopoietic development and functionally contributes to some of the SCL protein complexes. Finally, not only does SCL positively regulate expression of MYB, but it also prevents its degradation through miR-223-mediated repression of the tumor suppressor gene FBXW7.119 In conclusion, as in normal hematopoiesis, recursive circuits establish and maintain the oncogenic program controlled by SCL in T-ALL.
Toward new treatments of T-ALL?
ALL is a heterogeneous group of malignancies, covering a broad range of subtypes of B- and T-lymphocyte origin. The main therapeutic treatment of ALL relies on repeated cycles of chemotherapy, irrespective of the chromosomal abnormalities. This leads to ∼90% remission in children, but only 10% to 40% survival in adults due to toxicity and relapse.120 Dissecting the molecular mechanisms underlying the physiopathology of SCL+ T-ALL provides a basis for the development of novel therapies focusing on distinct aspects of SCL’s activities.
As oncogenic transformation relies on gene expression, pharmacological inhibition of components of the general transcriptional machinery has been explored. Low-dose inhibition of CDK7 kinase activity, necessary for phosphorylation of the C-terminal domain of RNA polymerase II, successfully reduced proliferation of an SCL+ T-ALL cell line.121 Importantly, the inhibitor predominantly affected expression of genes involved in the core regulatory circuitry (SCL, GATA3, RUNX1), possibly due to high sensitivity of the superenhancers regulating their expression.122 Epigenetic modifications have been the focus of a recent investigation. UTX, a histone H3K27 demethylase, is a member of the SCL complex in T-ALL that confers oncogenic properties.123 Remarkably, exposure of patient-derived xenotransplanted SCL+ T-ALL to H3K27 demethylase inhibitors selectively suppresses leukemic blast growth. Finally, the recent discovery that SCL regulates microRNA (miRNA) expression119,124 could pave the way to developing new therapeutic opportunities targeting miRNAs or their targets.
Protein/protein interactions (PPIs) are currently a major therapeutic focus. Although PPI interfaces are often large and featureless, making them difficult to target with small molecules, integration of structural, biochemical, and computational methods has opened the way to developing PPI inhibitors, providing a greater level of specificity.125 The x-ray structure of the SCL quaternary complex allows for the design of such inhibitory molecules.83 In particular, the small size of the SCL:LMO2 interface (620 Å2), together with the presence of defined secondary structures and identification of residues directly involved in SCL:LMO2 interaction, makes it an attractive target.83 As a prelude to these developments, both anti-LMO2 peptide aptamers and single-domain intracellular anti-LMO2 antibodies successfully inhibit LMO2-dependent T-cell tumor growth.126,127 Going forward, it will be essential to disrupt SCL-containing oncogenic complexes in a way that does not functionally affect the complexes required in normal hematopoiesis. Differences in affinity or stoichiometry between partners in distinct cellular contexts or tissue-specific PPIs would allow oncogenic-specific design or dosage of small-inhibitory molecules.
Conclusion
Recent years have seen significant advances in our understanding of SCL’s transcriptional mechanisms. The regulatory processes controlled by SCL are complex and many aspects remain to be investigated. In particular, the signaling pathways initiating cellular transitions during lineage development are not clear; the mechanistic relationships between SCL multiprotein complexes and chromatin remodelers need to be defined; the role of LYL1 should be further analyzed: redundancy with SCL in the HE could explain the apparent lack of SCL function in HSC emergence; and finally, functional correlation between SCL and miRNAs warrants further exploration. Ultimately, unraveling SCL-dependent normal and oncogenic processes may expose unsuspected lineage-specific pathways that will contribute to developing high-efficacy targeted therapies in SCL+ T-ALL.
Acknowledgments
The authors apologize to their colleagues whose work could not be cited due to space constraints. The authors thank Paresh Vyas, Claus Nerlov, and Aldo Ciau-Uitz for critical review of the manuscript.
The work performed in C.P.’s laboratory was supported by the Medical Research Council (MRC), and the Biotechnology and Biological Sciences Research Council (BBSRC).
Authorship
Contribution: C.P., H.C., and M.S.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine Porcher, Medical Research Council Molecular Haematology Unit, Medical Research Council Weatherall Institute of Molecular Medicine, John Radcliffe Hospital, Oxford OX3 9DS, United Kingdom; e-mail: catherine.porcher@imm.ox.ac.uk.