Abstract
The Runx family of transcription factors (Runx1, Runx2, and Runx3) are highly conserved and encode proteins involved in a variety of cell lineages, including blood and blood-related cell lineages, during developmental and adult stages of life. They perform activation and repressive functions in the regulation of gene expression. The requirement for Runx1 in the normal hematopoietic development and its dysregulation through chromosomal translocations and loss-of-function mutations as found in acute myeloid leukemias highlight the importance of this transcription factor in the healthy blood system. Whereas another review will focus on the role of Runx factors in leukemias, this review will provide an overview of the normal regulation and function of Runx factors in hematopoiesis and focus particularly on the biological effects of Runx1 in the generation of hematopoietic stem cells. We will present the current knowledge of the structure and regulatory features directing lineage-specific expression of Runx genes, the models of embryonic and adult hematopoietic development that provide information on their function, and some of the mechanisms by which they affect hematopoietic function.
Introduction
The family of Runx transcription factors (Runx1, Runx2, and Runx3) are involved in the regulation of lineage-specific genes, cell identities, and functions throughout development. Runx1 was first identified as a protein that binds in a sequence-specific manner to enhancers of Moloney murine leukemia virus1 and polyomavirus,2 and it was originally designated as core binding factor α (CBFα) and polyoma enhancer binding protein 2 (PEBP2), respectively. The gene encoding RUNX1 was cloned as one of the genes at the breakpoint of t(8,21) associated with acute myeloid leukemia, resulting in its original name AML1. Based on their homology to the Drosophila transcription factor runt, the name of this transcription factor family was changed to runt-related proteins or “Runx.”3 Each of the 3 mammalian Runx proteins contains a DNA-binding domain that is highly homologous to that of Drosophila runt. Runt domain–containing factors are found throughout the metazoan kingdom, from cnidarians and sponges to mammals, suggesting conserved roles for these factors in basic biological processes.4-6
In mammals, Runx1 was first identified as a critical regulator of hematopoiesis,7,8 Runx2 for its role in bone development,9,10 and Runx3 for its requirement in neurogenesis.11 However, all 3 factors were later shown to play critical roles also in other organ systems,12-14 and it is now clear that not only Runx1 but also Runx2 and Runx3 play a part in hematopoiesis. Cbfβ, the heterodimeric non–DNA-binding partner protein of the Runx factors, is also critical for hematopoiesis; without it, the Runx factors cannot bind DNA efficiently to regulate expression of their target genes.15
Within the hematopoietic system, Runx factors are expressed in a partially overlapping pattern. Runx activity as a transcription factor may be expected in all Runx-expressing cells, as its obligate partner, Cbfβ, is ubiquitously expressed.16,17 In this regard, the specific expression patterns of Runx genes predict which cell type will be affected by Runx deletion. However, where expression of the individual Runx genes overlaps, specific phenotypes may be absent due to redundancy.
The groundbreaking discovery of the requirement for Runx1 and Cbfβ in the embryonic generation of hematopoietic stem cells (HSCs) and the adult hematopoietic system7,8,15,18-21 and its role in normal adult hematopoiesis has fueled high interest and much research on this transcription factor, particularly in the regulation of clinically relevant HSCs. Here, we review the role of Runx transcription factors and Cbfβ in normal hematopoiesis, with a focus on HSCs and mammalian hematopoiesis. The involvement of Runx factors in immune cells is extensively reviewed elsewhere.22,23
Expression and the role of Runx factors in primitive hematopoiesis
Runx1 expression is dynamic during the time when the earliest blood cells are formed, with its expression slightly preceding the appearance of blood cells (Figure 1, left). In the yolk sac, the site of origin for the primitive hematopoietic lineages (ie, primitive erythrocytes, megakaryocytes, and macrophages), Runx1 is expressed in the mesoderm of the presumptive blood islands starting from E7.5 of mouse development.19 Expression continues upon blood island mesoderm differentiation into primitive erythrocytes, but it ceases shortly after the appearance of these cells.19 It was initially reported that primitive erythrocytes do not require Runx1 for their emergence, as they are present in normal numbers in Runx1-null embryos.7,15 This was surprising, given its expression during the generation of this lineage. More recently, it was reported that primitive erythrocytes do require Runx1 for their proper maturation. In its absence, they show morphological defects, with decreased expression of the erythroid marker Ter119 and the transcription factors EKLF and Gata1.24 This is a relatively mild phenotype compared with the dramatic effect that loss of Runx1 has on later hematopoietic cells of the embryo. It is unlikely that there is Runx factor redundancy at this stage. No or very low levels of Runx3 expression were detected in these cells,25 and an impaired maturation of primitive erythrocytes was also reported in embryos carrying a Cbfβ-MYH11 knockin allele.26 Of the other lineages, primitive megakaryocyte specification still occurs in Runx1-null embryos,27 while primitive macrophages are not formed in Runx1-null embryonic stem cell (ESC) differentiation cultures28 and are severely depleted in the yolk sac.29
Embryonic sites of Runx1 expression. Diagrams of the mouse conceptus at embryonic day 7.5 (E7.5), E8.5, and E10.5 showing representative Runx1 expression patterns. Runx1 expression (blue) is measured by β-galactosidase activity in histologic sections of Runx1LacZ/+ embryos19 in which the LacZ knockin results in a Runx1 knockout allele or by immunostaining with a Runx1-specific antibody. Hematopoietic sites and cells expressing Runx1 are shown below each embryo. Runx1 expression patterns were taken from previous studies.19,28,45,60,62,64,66,67 ab, allantoic bud; bi, blood islands; BL-CFC, blast colony-forming cells; c, chorion; pl and PL, placenta; vc, vessel of confluence; ys and YS, yolk sac.
Embryonic sites of Runx1 expression. Diagrams of the mouse conceptus at embryonic day 7.5 (E7.5), E8.5, and E10.5 showing representative Runx1 expression patterns. Runx1 expression (blue) is measured by β-galactosidase activity in histologic sections of Runx1LacZ/+ embryos19 in which the LacZ knockin results in a Runx1 knockout allele or by immunostaining with a Runx1-specific antibody. Hematopoietic sites and cells expressing Runx1 are shown below each embryo. Runx1 expression patterns were taken from previous studies.19,28,45,60,62,64,66,67 ab, allantoic bud; bi, blood islands; BL-CFC, blast colony-forming cells; c, chorion; pl and PL, placenta; vc, vessel of confluence; ys and YS, yolk sac.
Runx factors in yolk sac erythromyeloid and lymphoid progenitors
In contrast to the limited role for Runx1 and Cbfβ in primitive erythrocytes, both factors are critically required for the generation of all other hematopoietic cell types normally produced in the yolk sac beginning at E8 to E8.5.7,8,15,18,19,30,31 These are uni-, bi-, and multilineage hematopoietic progenitor cells (HPCs) with a generally limited lifespan,32 although specific cell types, such as erythromyeloid progenitor–derived tissue macrophages,33 B-1 B cells, and T-cell–restricted progenitors,34 can persist into adulthood. All these cell types are believed to derive from a special hemogenic subset of yolk sac endothelium in a process termed endothelial-to-hematopoietic transition (EHT).19,20,29,35,36 Yolk sac hemogenic endothelium expresses Runx119,37 (Figure 1, middle) and in the absence of Runx1 EHT is blocked, both in the embryo19,36 and in ESC differentiation cultures.38
RUNX factors in definitive hematopoiesis
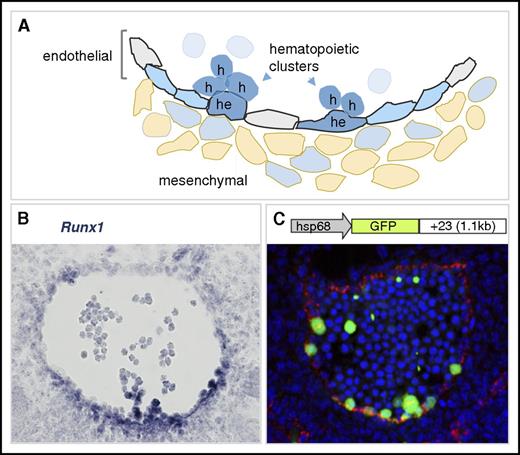
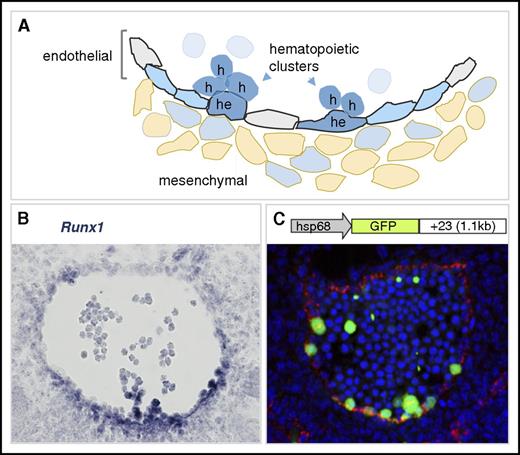
Definitive HSCs, the founders of the adult hematopoietic hierarchy, emerge first and autonomously in the major arteries (aorta, vitelline, and umbilical) of the mouse embryo. They are localized in the hematopoietic cell clusters that bud into the lumen of these vessels (Figure 1, right; and Figure 2A).39-43 The umbilical artery forms from the mesoderm in the allantois.44 Runx1 expression is seen from E7.5 in the allantois and from early E8 in endothelial cells of the nascent umbilical artery and the “vessel of confluence,” the point where the umbilical, yolk sac, and paired dorsal aortae amalgamate and from where the vitelline artery develops.45 In the mouse paired aortae, Runx1 expression starts from mid-E8 in a small subset of the endothelium, the hemogenic endothelium. Expression is initially ventral, but it later spreads to endothelial cells scattered around the entire circumference of the fused dorsal aorta and the emerging hematopoietic clusters.19,45,46 Runx1 is expressed in all emerging HPCs and HSCs in the vascular hematopoietic clusters (Figure 2A-B).19,39 Expression is also found in subaortic mesenchymal cells, starting from E10. These cells coexpress smooth muscle actin, suggesting they are developing smooth muscle cells.47,48 Finally, the early onset of Runx1 expression at all the sites of HSC emergence has implications for the interpretation of lineage-tracing experiments using tamoxifen-inducible Runx1-Cre mouse models.49,50 Together with the uncertainties about the perdurance of Cre nuclear localization, it is plausible that hemogenic endothelial cells at the sites of HSC emergence are labeled with early tamoxifen injections.
Runx1 expression in the mouse dorsal aorta during EHT. (A) Diagram of the ventral wall of the mouse E10.5 aorta showing Runx1-expressing cells (blue). Cells undergoing EHT are indicated as hemogenic endothelial (he) or hematopoietic cluster (h) cells. Aortic endothelial cells are indicated in gray, and Runx1-expressing hemogenic endothelial cells are in blue. Some mesenchymal cells (beige) underlying the aorta are Runx1 expressing. Runx1-expressing hematopoietic cell clusters are closely associated with the ventral luminal wall of the aorta. (B) Runx1 in situ hybridization analysis on a transverse section through the E10.5 mouse dorsal aorta. Runx1 expression (blue) is detected mainly at the ventral aspect of the aorta where it is seen in the subaortic mesenchymal cell layer, in some endothelial cells, and in hematopoietic cluster cells. In situ hybridization, microscopy, and image processing were performed as described in Bee et al.46 Original magnification ×400. (C) Schematic of the Runx1 +23 enhancer-reporter transgene construct (23GFP) and resulting GFP expression (green) in a transverse section through the E10.5 aorta of a 23GFP transgenic embryo. Endothelial cells (VE-cadherin labeled) are in red and nuclear stain (TO-PRO-3) in blue. The 23GFP transgene marks hemogenic endothelium in the vessel wall and all hematopoietic stem and progenitor cells (HSPCs) in the aortic clusters.55,135 Immunofluorescent labeling, confocal microscopy, and image processing were performed as in Swiers et al.55 Original magnification ×300.
Runx1 expression in the mouse dorsal aorta during EHT. (A) Diagram of the ventral wall of the mouse E10.5 aorta showing Runx1-expressing cells (blue). Cells undergoing EHT are indicated as hemogenic endothelial (he) or hematopoietic cluster (h) cells. Aortic endothelial cells are indicated in gray, and Runx1-expressing hemogenic endothelial cells are in blue. Some mesenchymal cells (beige) underlying the aorta are Runx1 expressing. Runx1-expressing hematopoietic cell clusters are closely associated with the ventral luminal wall of the aorta. (B) Runx1 in situ hybridization analysis on a transverse section through the E10.5 mouse dorsal aorta. Runx1 expression (blue) is detected mainly at the ventral aspect of the aorta where it is seen in the subaortic mesenchymal cell layer, in some endothelial cells, and in hematopoietic cluster cells. In situ hybridization, microscopy, and image processing were performed as described in Bee et al.46 Original magnification ×400. (C) Schematic of the Runx1 +23 enhancer-reporter transgene construct (23GFP) and resulting GFP expression (green) in a transverse section through the E10.5 aorta of a 23GFP transgenic embryo. Endothelial cells (VE-cadherin labeled) are in red and nuclear stain (TO-PRO-3) in blue. The 23GFP transgene marks hemogenic endothelium in the vessel wall and all hematopoietic stem and progenitor cells (HSPCs) in the aortic clusters.55,135 Immunofluorescent labeling, confocal microscopy, and image processing were performed as in Swiers et al.55 Original magnification ×300.
The spatiotemporal-restricted expression of Runx1 in the endothelial cells of the major embryonic arteries implicated a role for Runx1 in EHT in these locations. Indeed, in the absence of Runx1, no intra-aortic, vitelline, or umbilical artery hematopoietic cell clusters or functional HSPCs are detected.19,20,36,39 A similar requirement for Runx1 was seen in zebrafish EHT.51,52 Support for a role for Runx1 in hemogenic endothelium came from its conditional deletion in Tek- or vascular endothelial cadherin (VE-cadherin)–expressing cells. In these embryos, no hematopoietic cell clusters or functional HSCs or HPCs were formed, and embryos died around midgestation with fetal liver anemia, similar to the phenotype of Runx1-null embryos.20,35 These experiments did not distinguish between a role for Runx1 solely in the hemogenic endothelium or in the hematopoietic cluster cells, as VE-cadherin is expressed on the cluster cells and Cre could still be active there. However, Runx1 deletion in Vav-expressing hematopoietic cells did not phenocopy the Runx1 null, placing the limit of the critical Runx1 requirement prior to fetal liver colonization.20
Tober et al demonstrated that erythromyeloid progenitors and HSCs differ in their requirement for Runx1.53 Through inducible deletion of Runx1 in VE-cadherin+ cells in 24-hour intervals, they demonstrated that progenitors require Runx1 before E10.5, while the requirement for Runx1 in the generation of HSCs extended to E11.5. Interestingly, Liakhovitskaia et al54 reported that in the absence of Runx1, there were still some CD41+ cells in the wall of the E10.5 dorsal aorta with the phenotype of pre-HSCs, suggesting that initial hematopoietic specification does still occur. Conditional rescue of Runx1 in CD41-expressing cells restored functional HSC and HPC generation. At present, it is not clear whether Runx1 rescue occurred in CD41+ pre-HSCs54 or in endothelium of the early embryo, which was previously shown to express CD41.55 The endothelium to blood fate conversion was proposed to involve downregulation of Sox17, HoxA3, and Bmi1 that act upstream of Runx1.56-58 A recent review focuses on the essential role of Runx1 in the formation of blood cells from embryonic endothelium.59
Runx1 is also expressed in hematopoietic and endothelial cells of the placenta60,61 and in some cells of the embryonic head,62-64 2 other preliver sites where HSCs are found.60,65,66 The placenta is formed from a fusion of the chorion and allantois, both of which express Runx1 (Figure 1) and exhibit hematopoietic potential ex vivo.67 While the placenta generates multilineage progenitors de novo,61 it remains to be established whether HSCs are also generated in this tissue. In the human placenta, HSCs appear later than in the AGM, suggesting that this tissue is seeded by HSCs.68,69 HSC and HPC generation in head vasculature has proven elusive, with no evidence of EHT being reported.62-64
Interestingly, Runx1 haploinsufficiency during mouse development results in a dramatic change in the spatiotemporal distribution of HSCs, with a premature appearance of HSCs in the aorta and yolk sac.18 In mouse ESC differentiation cultures, Runx1 haploinsufficiency accelerates mesodermal development and hemangioblast specification.70 A gene regulatory network of a transcription factor triad (Scl, Gata2, and Fli1) that responds to Notch1 and BMP4 (through Runx1) signaling has been implicated by mathematical modeling to be involved in the accelerated emergence of HSCs when only 1 allele of Runx1 is expressed. In a simulation, the decrease in activation threshold in the haploinsufficient state results in an accelerated (∼2-day) triad activation.71
Only Runx1 plays a critical role at the onset of definitive hematopoiesis. Runx2 or Runx3 expression has not been reported in the aorta, vitelline, and umbilical arteries during EHT, and deletion of Runx29 or Runx372,73 did not result in the midgestation embryonic lethality and fetal liver anemia seen in Runx1-null embryos. In contrast, germline deletion of Cbfβ results in a phenotype similar to that of Runx1 germline deletion. Embryos die at E12.5 with fetal liver anemia and lack HSCs.15,30,31 Remarkably, Cbfβ-null fetal livers still contain a small number of HPCs, suggestive of a Cfbβ-independent role for Runx1 in early hematopoiesis, similar to that reported in zebrafish.74
RUNX factors in fetal and adult hematopoiesis
All blood cells in the adult (HSCs, HPCs, and mature blood cells) except erythrocytes express Runx1.75,76 MX-Cre conditional deletion of Runx1 in bone marrow (BM) chimeras showed an increase in lineage marker–negative, Sca1-positive, Kit-positive (LSK) cells, granulocytes, myeloid progenitors, and megakaryocyte progenitors, and T lymphocyte development was blocked.77-80 It was thought that Runx1 plays a role in regulating homeostatic HSC and HPC numbers in the BM. Although discrepancies in the phenotypic marker expression on hematopoietic cells were found in the absence of Runx1,77 long-term reconstituting (LTR) HSC frequency was three- to fourfold decreased as determined by peripheral blood chimerism. However, there was no change in BM chimerism, and thus, it was concluded that there is minimal impact of Runx1 on adult BM LTR HSCs.
Vav-Cre conditional deletion was used to examine whether HSCs are dependent on Runx1 and Cbfβ after they are generated. The onset of Vav-Cre activity is at E11.5 in HSCs.20 Whereas the deletion of Runx1 showed no significant reductions in phenotypic HSC or LTR HSCs in the E14.5 fetal liver,77 CBFβ deletion severely compromises LTR HSCs.53 Thus, Runx2 and/or Runx3 are likely able to contribute to the maintenance of HSCs. These results, together with the fact that Runx3 is expressed in fetal liver hematopoietic precursors and partially overlaps with Runx1 expression,81 support the notion that Runx family proteins act in a redundant manner in HSCs after they are generated.
Interestingly, the double deletion of Runx1 and Runx3 (MX-Cre) in adult mice was found to lead to BM failure (Fanconi anemia–like) and myeloproliferative disease.82 BM failure was found to be the result of differentiation blocks in all hematopoietic lineages and progressive HSC exhaustion. DNA repair was found to be defective, and it was concluded that repair relies on Runx-dependent recruitment of Fanconi proteins to the site of DNA damage. This activity is independent of Cbfβ, suggesting that Runx1 and Runx3 may function here in a non–transcription-related manner.
Haploinsufficiency of Runx1 also affects HSCs in the adult BM. LTR-HSCs are decreased by 50%, and upon transplantation, these HSCs provided enhanced engraftment and normal numbers of most hematopoietic lineages as compared with wild-type mice. In contrast, a decrease in CD4+ T cells and circulating platelets was observed.83 Runx1 haploinsufficient myeloid progenitors were found to be differentiation impaired. Although granulocyte lineage commitment occurs, progenitors do not mature84 and are hypersensitive to granulocyte colony-stimulating factor, predisposing the progenitors to expansion and mobilization rather than differentiation.85 Similarly, in humans, autosomal-dominant mutations in 1 Runx1 allele result in qualitative and quantitative platelet defects and a propensity to develop acute myeloid leukemia.86 Most recently, Vav-Cre–mediated deletion of Runx1 has been found to result in an increase in the stress resistance of BM HSCs and the superior survival of HSCs in the presence of genotoxic shock. Runx1 binds to promoters of genes encoding ribosome proteins, and in its absence, ribosome biogenesis is reduced,87 suggesting that this condition may facilitate the premalignant cell state.
Runx1 and Cbfβ have been found to also play a role in the regulation of pro–B cells and double-negative thymocytes, and the compound disruption of Runx1 and Runx3 genes suggests partially redundant function.88-90 Runx1 is expressed in earliest developing thymocytes and later in cortical thymocytes at E18 and postnatal week 6.90 Its expression is highest in the most immature thymocytes,91 supporting a role in T-cell lineage fate decisions. In the adult, Foxp3+ regulatory T cells and Th17 cells73 express Runx1. It is also expressed in Flt3+ dendritic cell precursors and common lymphoid progenitors, and Cbfβ is critical for the differentiation of these cells.92
Runx3 plays a role in innate immune cell types and T lymphoid development. Runx3-deficient mice exhibit defects in splenic natural killer cells, lack Langerhans and dendritic epithelial cells,72,73 and the lineage commitment of innate lymphoid cells is affected.93 At E18, Runx3 is expressed in thymocytes (medulla and cortical) and in the adult it is detected mainly in the medulla.90 Runx3 null mice exhibit defects in the differentiation of CD8+ cytotoxic T cells and Th1 cells.72,73 It is most highly expressed in mature thymocytes91 and is thought to act together with Runx1 as a transcriptional repressor of CD4 during T-cell lineage decisions.
Although Runx2 expression is high in the HSPC compartment of mouse BM,94 there is no data thus far demonstrating a role for Runx2 in fetal or adult BM HSCs.
Runx1 mechanism of action
Given the critical role of Runx1 in the establishment and maintenance of the adult hematopoietic system, there has been much interest in elucidating how it exerts it role. Runx1 acts as an epigenetic regulator and as an activator or repressor of transcriptional programs (reviewed in Tober et al,59 Lausen,95 Lichtinger et al,96 Obier and Bonifer,97 and Swiers et al98). In the dorsal aorta, Runx1 drives hematopoietic specification, which coincides with loss of endothelial potential.55-58 In ESC hematopoietic differentiation cultures, Runx1 is involved in the initial remodeling of the Pu.1 transcription factor locus, and Pu.1 is one of its important downstream targets.99,100 Intriguingly, this required only low-level transient expression of Runx1. Genome-wide analysis showed that in hemogenic endothelium, binding of SCL, Fli1, and Cebpβ was associated with the priming of a large number of hematopoietic loci. Upon upregulation of Runx1, these complexes relocated to new cis elements, now bound by Runx1 and showing increased histone acetylation. Thus, Runx1 changes the epigenetic landscape at hematopoietic loci to promote hematopoietic differentiation.101
Runx1 expression in ESC-derived hemogenic endothelium induces adhesion and migration-associated transcriptional profiles before the downregulation of endothelial genes.102 The Runx1 targets Gfi1 and Gfi1b are implicated in the downregulation of the endothelial program.103 They do this through recruitment of the chromatin modifier LSD1 (a member of the CoREST repressive complex) to endothelial gene loci.104 Later in hematopoietic differentiation, Runx1 positively regulates the expression of a large number of hematopoietic lineage–specific genes, including cytokines, cytokine receptors, and T-cell receptors,23,59,98 many of which are effectors of hematopoietic fate, differentiation, and function. Whether Runx1 functions as a transcriptional activator or repressor is dependent upon cofactor interactions. Posttranslational modifications by tyrosyl phosphorylation regulate the interactions of Runx1 with different coregulators.105
Structure of mammalian Runx genes and alternatively spliced isoforms
The genomic organization of mouse and human Runx genes is highly conserved. All vertebrate Runx genes have 2 promoters (distal P1 and proximal P2) and a variable number of alternatively spliced protein-coding exons (Figure 3).4,5,106-110 P1- and P2-derived alternative transcripts are differentially expressed in various cell types and developmental stages. Below, we focus on Runx1 as the main regulator of hematopoiesis, but also Runx2 and Runx3 express transcripts from 2 promoters to generate alternative isoforms.94,111-113
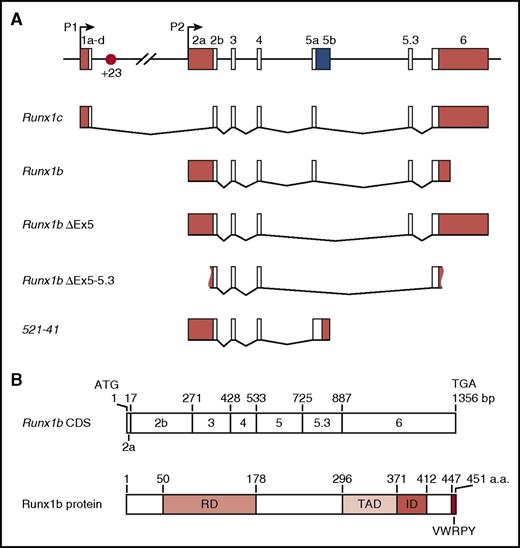
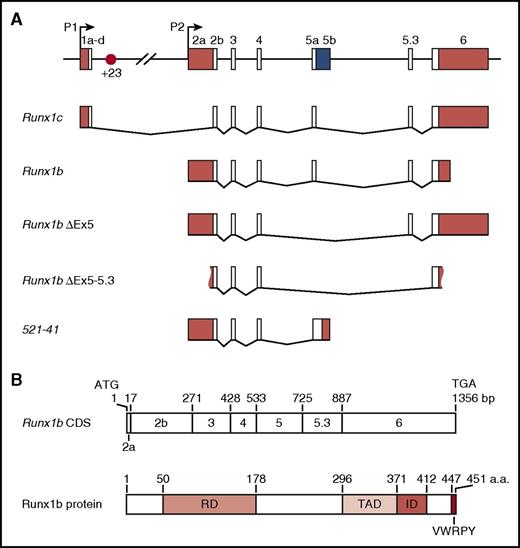
Runx1 locus and isoforms. (A) Schematic of the 224-kb mouse Runx1 locus, alternative splice forms, and the +23 enhancer (not to scale) reported in the literature.46,108,109,114,128,142,143 White boxes, coding sequence; red boxes, untranslated regions (UTRs); blue box, predicted coding +UTR. Figure 3A reprinted with minimal alteration from Bee et al46 with permission from Elsevier. (B) Schematic showing Runx1 protein domains relative to their coding exons.110 CDS, coding sequence; ID, inhibitory domain; RD, runt domain; TAD, transactivation domain.
Runx1 locus and isoforms. (A) Schematic of the 224-kb mouse Runx1 locus, alternative splice forms, and the +23 enhancer (not to scale) reported in the literature.46,108,109,114,128,142,143 White boxes, coding sequence; red boxes, untranslated regions (UTRs); blue box, predicted coding +UTR. Figure 3A reprinted with minimal alteration from Bee et al46 with permission from Elsevier. (B) Schematic showing Runx1 protein domains relative to their coding exons.110 CDS, coding sequence; ID, inhibitory domain; RD, runt domain; TAD, transactivation domain.
Alternative promotor usage contributes to regulatory complexity, with the production of Runx1 transcripts that differ in their 5′ and 3′ UTRs, their coding sequence, and the time required for their transcription (Figure 3).46,81,106,114 Differences in the identity and length of the UTRs have been linked to differences in RNA stability and translation efficiency.115,116 P1-derived transcripts are subject to Cap-mediated translation, while P2-derived transcripts undergo internal ribosome entry site–mediated translation. The latter is thought to facilitate translation under conditions of stress and during cell division.116 Translation of Runx1 is also subject to regulation by microRNAs.117,118 Several alternatively spliced transcripts have been described in mouse (Figure 3) and human,119 including the full-length P1- and P2-derived Runx1c and Runx1b. One of these transcripts, RUNX1a, lacks the transactivation domain while retaining the DNA-binding domain and has been proposed to act as a dominant negative.119,120
The biological relevance of the different Runx1 isoforms is not fully understood. Exclusive expression of the full-length Runx1 is sufficient for in vitro hematopoiesis and for embryos to develop into adulthood,28,75,121,122 and both the runt domain and the transactivation domain are essential.121,122 As no detailed functional analyses of HSCs and HPCs in vivo were performed, the precise role of this and other Runx1 isoforms in HSC biology remained unclear. More recently, enforced expression of the short human RUNX1a isoform was shown to favor expansion of the HSC pool, in line with earlier reports that it supports proliferation in a myeloid cell line.123-126 Expression of the full-length RUNX1b and RUNX1c, in contrast, promoted differentiation, depleting the HSC pool. A similar short isoform lacking the transactivation domain was described in mice, albeit with a different 3′ exon (Figure 3).127,128 Interestingly, the mouse short Runx1 isoform had a biological role similar to that of the human RUNX1a in that it positively regulated the size of the HSC pool.127
The full-length P1- and P2-derived Runx1c and Runx1b protein isoforms are identical apart from their N terminus. In human and mouse, there are 32 and 19 unique RUNX1c N-terminal amino acids, respectively, and 5 unique RUNX1b ones. In the mouse, no differences in hematopoietic development were observed in ESC differentiation cultures upon overexpression of Runx1c or Runx1b.129 In human, however, a recent study reported on a specific role for the RUNX1c in regulating the growth of Epstein-Barr virus–transformed B cells, unearthing a first indication for specific functionality associated with this domain. Interestingly, the sequence responsible for this effect is specific to the human RUNX1c N terminus.130
Tissue-specific expression of RUNX
All 3 Runx proteins bind the same 5′-ACCG/ACA-3′ DNA motif and their specific roles are due to their spatiotemporal expression patterns. Both promoters of human and mouse RUNX1 contain consensus binding motifs for several hematopoietic transcription factors (E-box, Ets, and Runx), but these are not sufficient to mediate tissue-specific transgene expression.106,131 The P2 promoter lies in a large CpG island, often found at promoters of broadly expressed genes. The P1 promoter, which shows greater tissue-specific activity than P2,107 contains 2 adjacent conserved Runx motifs just downstream of the transcription start site (conserved in all 3 Runx genes). These play a role in autoregulation of Runx1 during hematopoietic development and in auto- and crossregulation in the adult (reviewed in Bee et al46 ). Interestingly, Smad1, a positive mediator of Bmp4 signaling, was found to transactivate the Runx1 P1 promoter, whereas Smad6 acts as an inhibitor.132 Also, the Wnt pathway was reported to activate the RUNX1 P1 promoter.133
Runx1 promoters show specific activity patterns during developmental differentiation of cells to the hematopoietic lineage. At all sites of HSPC emergence, P2 promoter activity precedes that of P1.46,129,134,135 The P1 promoter is activated when hemogenic cells commit to a hematopoietic fate. As hematopoietic development proceeds in the fetal liver, P1 becomes the dominant promoter and remains so in adult BM.46 A similar switch in promoter usage is seen in the developing thymus.136 Mouse and human ESCs undergoing hematopoietic differentiation also show P2 promoter usage before P1.126,134,137,138 The sequential P2 to P1 promoter usage may reflect a more ancient role for P2 (present in all metazoan Runx genes) than P1, which is vertebrate specific.4,5
Although the Runx1 promoters do not contribute tissue specificity, they play important, nonredundant roles in hematopoiesis. Analysis of a P1-null mouse model showed that lack of Runx1c negatively affects the initial emergence of HPCs in the embryo, but embryos survive into adulthood.135 Adult P1-null mice show a phenotype in between Runx1 haploinsufficient and Runx1 MX-CRE–deleted mice.135 Loss of P1 also leads to severely reduced numbers of basophils and T-cell lineage defects.139,140 In an attenuated P2 promoter mouse model, embryos die around midgestation with hemorrhages, fetal liver anemia, and defects in thymocyte development.136 The generation of hematopoietic progenitors and aortic cell clusters is severely affected,135 and this phenotype is in line with the earlier and broader expression of P2-derived isoforms. Thus, both Runx1 promoters contribute to the expression of normal spatiotemporal levels of Runx1. How alternative promoter expression is regulated is poorly understood. A study in zebrafish suggests a role for cohesion and CTCF.141
The finding that Runx1 promoters do not direct tissue-specific expression prompted the search for enhancers that mediate Runx1 expression during EHT and in HSCs. Using a combination of comparative genomics, chromatin accessibility assays, and transient mouse transgenesis, a highly conserved hematopoietic Runx1 +23 enhancer was identified in the first intron of the 224-kb mouse gene.142 It was shown that the +23 enhancer is sufficient to mediate reporter gene expression in a pattern similar to Runx1 during hematopoietic development. It directs reporter gene expression in all emerging HSCs and HPCs and labels all hematopoietic-fated endothelial cells in the mouse embryo (Figure 2C).55,135,142,143 Of note, it does not mark the subaortic Runx1+ smooth muscle cells or any other nonhematopoietic Runx1-expressing tissues in the embryo.55,142 The mouse +23 enhancer was reported to also mark hematopoietic-fated endothelial cells and hematopoietic cells in zebrafish,143-145 although it is not conserved in this species.131 In human ESC and induced pluripotent stem cell differentiation cultures, the +23 enhancer in conjunction with the RUNX1 P1 promoter is active in hematopoietic cells and in colony-forming units in culture.146 Thus, the Runx1 +23 enhancer is a valuable tool in studies examining hematopoietic development.
Insight into the pathways converging on Runx1 will yield important information on signals underlying hematopoietic specification. A recent review59 summarizes the known transcription factors and pathways acting upstream of Runx1. Some of these act directly on the Runx1 +23 enhancer. Indeed, +23 enhancer activity is dependent on Gata, Runx, and Ets motifs; Gata2, Ets factors (Fli-1, Elf-1, and Pu.1), and the SCL/Lmo2/Ldb1 complex are bound to the enhancer in vivo in yolk sac and fetal liver.25,142 In addition to the +23 enhancer, several other hematopoietic Runx1 enhancers also mediate reporter gene expression to the dorsal aorta and fetal liver in a Runx-specific pattern.147 Most of these also contain binding motifs for Gata, Runx, and Ets and are bound by these and other hematopoietic transcription factors. The precise regulatory roles of these enhancers on Runx1 expression in developmental hematopoiesis, and the developmental pathways converging on them, remain to be established.
Implications and perspectives
Several decades of research on the Runx protein family has provided a wealth of molecular and biological information concerning their roles in normal hematopoiesis. Their many isoforms and expression patterns highlight a complexity that goes beyond a simple description of 3 transcription factors. The overlapping expression of Runx1 and Runx3 in some hematopoietic cell lineages reveals functional redundancy. As all 3 Runx proteins interact with their obligate partner, Cbfβ, deletion of Cbfβ has more profound consequences for hematopoietic cells than single Runx gene deletions. Novel mouse models reveal the mechanisms underlying Runx function, such as in ribosome biosynthesis, which in Runx1 deficiency affects the stress status of HSCs. These long-term in vivo studies are for the first time providing an understanding of the preleukemic condition that affects the balance of Runx-centric gene regulatory networks and epigenetic status. Moreover, roles for Runx proteins outside of their classification as transcription factors are being proposed, such as in DNA repair in which Runx acts independently of Cbfβ.
As the Runx field moves forward, new methods of single-cell analysis are being employed. Time-lapse images of single Runx1-expressing hematopoietic cells during emergence from hemogenic endothelium confirmed EHT and in the future are likely to provide important information on the dynamic expression of Runx1, transcription factor assemblies, and reshaping of the epigenetic landscape during this process. Transcriptomic and proteomic signatures are expected to elucidate the inherent instability of hematopoietic cells and their heterogeneity during hematopoietic cell fate transitions in development and adult hematopoietic differentiation and during aging. Together, this and future knowledge on Runx proteins will benefit biomedical approaches for the treatment of Runx-related hematologic dysfunctions and pathologies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Chris S. Vink and Claudia Baumann for the in situ hybridization and immunostained images in Figure 2.
M.d.B. is supported by an MRC Molecular Hematology Unit (Oxford) Core Award. E.D. is supported by ZonMW TOP (91211068) and a European Research Council Advanced Grant (341096).
Authorship
Contribution: M.d.B. and E.D. reviewed the literature and wrote the review.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elaine Dzierzak, University of Edinburgh, Centre for Inflammation Research, Queens Medical Research Institute, 47 Little France Crescent, Edinburgh, EH16 4TJ, United Kingdom; e-mail: elaine.dzierzak@ed.ac.uk.