Key Points
Individual histone proteins and DNA purified from normal human neutrophils promote coagulation activation.
Neither intact NETs nor nucleosomes directly promote coagulation activation in plasma in vitro.
Abstract
NETosis is a physiologic process in which neutrophils release their nuclear material in the form of neutrophil extracellular traps (NETs). NETs have been reported to directly promote thrombosis in animal models. Although the effects of purified NET components including DNA, histone proteins, and neutrophil enzymes on coagulation have been characterized, the mechanism by which intact NETs promote thrombosis is largely unknown. In this study, human neutrophils were stimulated to produce NETs in platelet-free plasma (PFP) or in buffer using phorbol myristate actetate or calcium ionophore. DNA and histone proteins were also separately purified from normal human neutrophils and used to reconstitute chromatin using a salt-gradient dialysis method. Neutrophil stimulation resulted in robust NET release. In recalcified PFP, purified DNA triggered contact-dependent thrombin generation (TG) and amplified TG initiated by low concentrations of tissue factor. Similarly, in a buffer milieu, DNA initiated the contact pathway and amplified thrombin-dependent factor XI activation. Recombinant human histones H3 and H4 triggered TG in recalcified human plasma in a platelet-dependent manner. In contrast, neither intact NETs, reconstituted chromatin, individual nucleosome particles, nor octameric core histones reproduced any of these procoagulant effects. We conclude that unlike DNA or individual histone proteins, human intact NETs do not directly initiate or amplify coagulation in vitro. This difference is likely explained by the complex histone-histone and histone-DNA interactions within the nucleosome unit and higher-order supercoiled chromatin leading to neutralization of the negative charges on polyanionic DNA and modification of the binding properties of individual histone proteins.
Introduction
NETosis is a recently described process in which nuclear material in the form of a meshwork of chromatin is released by neutrophils into the extracellular space. These structures are known as neutrophil extracellular traps (NETs).1 NETs are composed of a backbone of DNA coiled around histones and decorated with enzymes derived from neutrophil cytoplasmic granules. NETosis is thought to be an antimicrobial defense mechanism used by neutrophils.1
Experimental studies in animal models have shown that NETs also promote thrombosis.2-4 Their dual role in immune defense and thrombosis has led to the proposal that NETs are a major player in “immunothrombosis,” in which immune system effectors cooperate with the clotting system to delimit infection at the expense of thrombosis.5 Several studies have used purified NET components to investigate the mechanism(s) by which NETs promote thrombosis. Specifically, neutrophil elastase has been shown to inactivate tissue factor (TF) pathway inhibitor.6 Purified histones enhance thrombin generation (TG) in plasma through platelet-dependent7 and platelet-independent mechanisms.8 They also enhance polyphosphate-triggered TG in plasma7 and promote phosphatidylserine exposure on red blood cells.9 Purified DNA and purified RNA have been shown to bind and activate proteins of the contact system in buffer and enhance TG and clot formation in platelet-poor plasma (PPP).10-12 Blood perfused over intact NETs leads to clot formation, where platelets are recruited and activated through the contribution of NET-bound von Willebrand factor.3 TF expression on NETs released in the context of autoimmune vasculitis has been reported.13 Finally, in the presence of intact NETs, TG was enhanced via contact activation in PPP and through histone-mediated platelet activation in platelet-rich plasma (PRP).14 However, the conditions used to prepare PPP (by single centrifugation of whole blood at 1500g for 10 minutes) may have contained substantial numbers of residual platelets.15 Therefore, it remains unclear whether the enhanced TG observed was a direct effect of intact NETs on coagulation or was influenced by contaminating platelets.
In this report, neutrophils were isolated from whole blood with minimal platelet contamination to compare the effects of purified human neutrophil DNA (hnDNA) or human histone proteins with those of reconstituted nucleosomes, chromatin, and intact NETs on coagulation in vitro.
Methods
Materials
All buffers, salts, and bovine serum albumin (BSA) were of high-quality reagent grade and were from ThermoFisher Scientific (Waltham, MA). Unilamellar phospholipid vesicles (15% phosphatidylserine [PS], 41% phosphatidylcholine [PC], and 44% phosphatidylethanolamine [PE]) were prepared as described.16 Factor XII (FXII), FXI, FXIa, antithrombin (AT), and recombinant tissue factor (TF1-263) were from Haematologic Technologies Inc. (Essex Junction, VT). FXIIa, high-molecular-weight kininogen (HMWK), prekallikrein, and recombinant hirudin were from Enzyme Research Laboratories (South Bend, IN). TF was relipidated in PCPS (25% PS, 75% PC) vesicles as previously described.17,18 Corn trypsin inhibitor (CTI) was prepared as previously described.19 Sytox Green Nucleic Acid Stain, DNAase I, streptavidin–horseradish peroxidase, and 5-thio-2-nitrobenzoic acid–peroxidase substrate and the S-2366 substrate were from ThermoFisher Scientific (Waltham, MA). Phorbol myristate actetate (PMA), calf thymus histones, A23187, and 0.4% trypan blue solution were from Sigma-Aldrich (St. Louis, MO). The fluorogenic substrate Z-GGR-AMC was from Bachem (Bubendorf, Switzerland). Human XII-, XI-, and VII-deficient plasmas were from George King Bio-medical Inc. (Overland Park, KS). Recombinant human histones H3 and H4 were from New England Biolabs (Ipswich, MA); TRAP6 was from R&D Systems (Minneapolis, MN). The primary rabbit polyclonal antibody against citrullinated histone H3 was from Abcam (Cambridge, MA), whereas the DyLight 594 Goat Anti-Rabbit IgG Antibody was from Vector Laboratories (Burlingame, CA). Recombinant human histone octamers and nucleosomes were from EpiCypher (Chapel Hill, NC).
Neutrophil isolation and stimulation for NET formation
Neutrophils were isolated from whole blood by a magnetic bead–based negative selection using the MACSxpress Neutrophil Extraction Kit (Miltenyi Biotec, San Diego, CA) and a strong magnet (MACSxpress Separator, Miltenyi Biotec). Residual red blood cells were removed by repeat negative selection after incubation of resuspended neutrophils with magnetic beads coated with antibodies against glycophorin A (Miltenyi Biotec) and flowing through a porous column (25 LD MACS separation column, Miltenyi Biotec) placed in the magnetic field (MidiMACS, Miltenyi Biotec). Harvested neutrophils were washed twice with RPMI and resuspended either in plasma (PRP or platelet-free plasma [PFP]) for TG assay or in RPMI 1640 + 1% BSA + 2 mM calcium + 1.6 mM magnesium for the synthetic contact system assay (SCSA) as described subsequently. The purity of the final neutrophil suspension assessed using a Sysmex pocH-100i Hematology Analyzer (Sysmex, Kobe, Japan) was ≥97%, and viability (by trypan blue exclusion) was ≥98%.
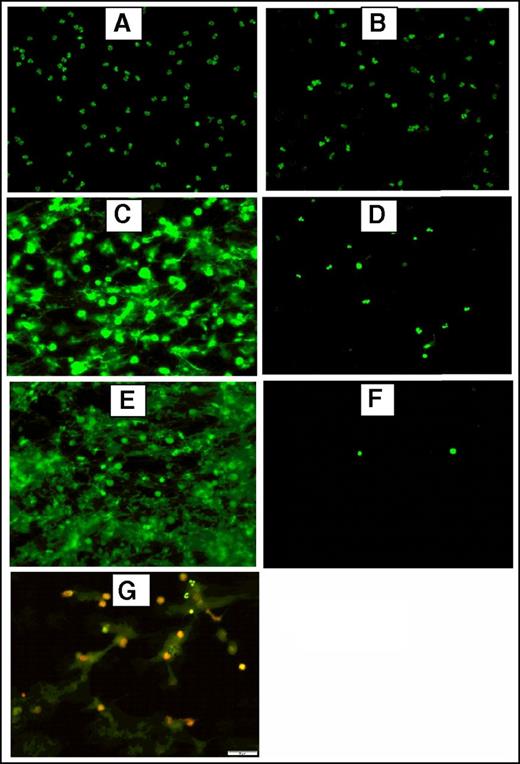
To generate NETs, resuspended neutrophils were adjusted to 2500/µL, 5000/µL or 12 000/µL and stimulated with 600 nM PMA or 5 µM ionophore at 37°C and 5% CO2 for 3 hours. Fluorescence and immunofluorescence microscopy were used to verify NET formation after stimulation and incubation of 5 × 105 neutrophils seeded on a slide coated with poly-l-lysine as described previously (Figure 1). For immunofluorescence microscopy, samples were incubated overnight at 4°C with a primary rabbit antibody against citrullinated histone H3 (Abcam) (1:250). A secondary DyLight 594 goat anti-rabbit IgG antibody (Vector Laboratories, Burlingame, CA) (1:200) was incubated 1 hour at room temperature in the dark. The preparations were washed 3× with 1× PBS after each incubation step. Prepared slides were analyzed using the ×20 objective lens of an Olympus BX51WI microscope equipped with an Olympus DP80 camera using the CellSens software.
Fluorescence and immunofluorescence microscopy of stimulated human neutrophils for NETs formation. Stimulated normal human neutrophils were seeded on poly-l-lysine–coated slides and incubated at 37°C + 5% CO2 for 3 hours. After incubation, the preparations were fixed using 4% paraformaldehyde and stained using 2.5 μM Sytox Green, a DNA staining dye. Unstimulated neutrophils did not form NETs (A) and were intact after treatment with DNAase (20 μM) (B). Robust NET release after stimulation with 600 nM PMA (C) or 5 μM A23187 (E) as shown by the bright and scattered Sytox Green signal pattern. Digestion of PMA-stimulated (D) and A23187-stimulated (F) NETs after treatment with DNAase (20 μg/mL), indicating the extracellular location of NETs. (G) Dual immunofluorescence staining of NETs by Sytox Green (2.5 μM) for DNA and citrillunated histone H3 (red). A23187, calcium ionophore.
Fluorescence and immunofluorescence microscopy of stimulated human neutrophils for NETs formation. Stimulated normal human neutrophils were seeded on poly-l-lysine–coated slides and incubated at 37°C + 5% CO2 for 3 hours. After incubation, the preparations were fixed using 4% paraformaldehyde and stained using 2.5 μM Sytox Green, a DNA staining dye. Unstimulated neutrophils did not form NETs (A) and were intact after treatment with DNAase (20 μM) (B). Robust NET release after stimulation with 600 nM PMA (C) or 5 μM A23187 (E) as shown by the bright and scattered Sytox Green signal pattern. Digestion of PMA-stimulated (D) and A23187-stimulated (F) NETs after treatment with DNAase (20 μg/mL), indicating the extracellular location of NETs. (G) Dual immunofluorescence staining of NETs by Sytox Green (2.5 μM) for DNA and citrillunated histone H3 (red). A23187, calcium ionophore.
Neutrophils resuspended in RPMI were also stimulated as described previously for NETs formation in cell culture dishes. After discarding the supernatant, NETs were harvested in 5 mL of new medium and centrifuged at 300g for 10 minutes to pellet intact cells. Then, the supernatant was further centrifuged at 20 000g for 30 minutes to pellet NETs. Washed NETs were then resuspended in 1 mL of RPMI 1640 + 1% BSA. Nucleosomes and cell-free DNA were measured in washed NET preparations to confirm the presence of NETs.
Plasma preparation
Whole blood was collected in plastic tubes containing 3.2% citrate at a ratio of 1 volume citrate to 9 volumes blood, from adult healthy medication-free volunteers, under an approved human subjects protocol. PRP was prepared by centrifugation at 200g for 10 minutes. The supernatant was carefully collected and adjusted with autologous PFP to a final platelet concentration of 200 × 109/L. PFP was prepared by double centrifugation at 2500g for 15 minutes and filtration through a 0.22-µm filter (ThermoFisher Scientific).
TG assay
TG assays were performed as previously described,20 with some modifications. Briefly, 80 µL of PFP or PRP, containing hnDNA, histones, nucleosomes, reconstituted chromatin, or NETs was placed in wells of a black 96-well microplate. Plasma was recalcified (15 mM CaCl2) for 3 minutes in the presence of 416 μM Z-GGR-AMC fluorogenic substrate prior to activation with 4 μM PCPEPS alone or with 1 pM recombinant TF and 4 μM PCPEPS. No synthetic phospholipids were added in PRP. Substrate hydrolysis was monitored in a Biotek Synergy H1M fluorometer, and TG was calculated based on a thrombin standard curve.
SCSA
The proteins of the contact pathway (FXII, prekallikrein, HMWK, and FXI) were reconstituted in buffer (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid buffered saline, 0.1% polyethylene glycol [PEG] 800, 2 mM Ca2+, 1.6 mM Mg2+, 20 µM Zn2+) or RPMI 1640 + glutamate and 1% BSA, 2 mM Ca2+, 1.6 mM Mg2+, and 20 µM Zn2+ at physiologic concentrations (375 nM, 580 nM, 670 nM, and 30 nM, respectively). FXII activation was initiated by the addition of kaolin (positive control), purified genomic hnDNA, and quiescent or stimulated neutrophils. Aliquots were removed and quenched with an equal volume of buffer containing 7 µM AT, 0.2 mg/mL CTI, and 10 U/mL unfractionated heparin. Samples were stored at 4°C prior to quantification of FXIa-AT complexes using an in-house enzyme-linked immunosorbent assay (ELISA).
FXIa-AT ELISA
The 96-well microplates were coated with 5 µg/mL mouse anti-human FXI antibody overnight at 4°C. Plates were washed 3× with wash buffer (Hepes-buffered saline [HBS], 1% BSA, 0.05% Tween 20) and blocked with HBS + 2% milk for 1 hour at room temperature, before being subjected to 3 further washes. A FXIa-AT standard was constructed by reacting human FXIa (1 µM) with human AT (3 µM) in the presence of unfractionated heparin (10 U/mL) for 30 minutes at 37°C. Samples and standards were incubated on a rocker for 1 hour at room temperature. Plates were then washed 3×, and 5 µg/mL biotinylated burro anti-human AT was added for 1 hour at room temperature. Plates were then washed 3×, streptavidin–horseradish peroxidase (1:10 000) was added, followed by incubation for an additional 30 minutes. The wells were washed 3× and then incubated with a 5-thio-2-nitrobenzoic acid–peroxidase substrate prior to quenching with 2M sulfuric acid. Plates were read on a Molecular Devices Spectramax microplate reader.
Chromatin reconstitution
Histones and DNA were purified from normal human neutrophils using Histone Extraction Kit (Abcam) and QIAamp DNA Blood Midi Kit (Qiagen GmbH, Hilden, Germany), respectively, according to the manufacturers’ instructions. Purified DNA and histones from human neutrophils were then used for chromatin reconstitution by salt-gradient dialysis as previously described.21 Briefly, histones and hnDNA were mixed together in a solution of 1M NaCl, 1 mM EDTA at pH 8, with a 1:1.2 mass ratio. Purified histones and hnDNA were also diluted at the same concentration in the same buffer separately. One milliliter of each of the 3 solutions was introduced into Slide-A-Lyzer Dialysis cassettes (ThermoFisher Scientific) equipped with 7000 molecular weight cut-off dialysis membrane. The cassettes filled with solutions were placed successively in 3 solutions of 0.8, 0.15, and 0 M NaCl for 2 hours, 2 hours, and overnight, respectively. All 3 solutions contained 20 mM tris(hydroxymethyl)aminomethane-HCl and 0.2 mM Na2EDTA at pH 7.2. After dialysis, the samples were concentrated using 10 kDa molecular weight Amicon Ultra centrifugal filter units (EMD Millipore, Darmstadt, Germany) spun at 3000g for 15 minutes.
Nucleosome measurement
Nucleosomes were quantified using the Cell Death Detection ELISAplus kit (Roche Diagnostics Corporation, Indianapolis, IN), according to the manufacturer’s instructions.
Chromogenic assays
Contact activation assay.
FXII, HMWK, and FXI were diluted in a buffered system (HBS, 0.1% PEG 800, 2 mM Ca2+, 1.6 mM Mg2+, 20 µM Zn2+) at physiologic concentrations. The contact system was then activated using kaolin (positive control), buffer (negative control), hnDNA, purified neutrophil histones, or reconstituted chromatin. After incubation at 37°C for 20 minutes, the reaction was quenched with CTI (0.1 mg/mL). Then, a chromogenic substrate for FXIa (S-2366) was added at 200 mM final concentration, and the microplate was read immediately in the kinetic mode at 405 nm for 2 minutes.
Thrombin-dependent FXIa generation assay.
FXI and HMWK were reconstituted in buffer (HBS, 0.1% PEG 800, 2 mM Ca2+, 1.6 mM Mg2+, 20 µM Zn2+) at physiologic concentrations (Zymogen solution). Then 40 µL was mixed with 10 µL of α-thrombin (10 nM final concentration), in the absence or presence of hnDNA, purified neutrophil histones, or reconstituted chromatin. FXIIa (25 nM) was used as positive control. All samples were incubated at 37°C for 20 minutes, after which the reaction was quenched with recombinant hirudin (100 nM final concentration). A chromogenic substrate for FXIa (S-2366) was then added at 200 mM final concentration, and the microplate was read immediately in the kinetic mode at 405 nm for 2 minutes.
Statistical analyses
All the variables compared showed a normal distribution using the Kolmogorov-Smirnov test. The t test was used to compare between 2 conditions, whereas the analysis of variance test with Tukey’s multiple correction test was used to compare between >2 conditions. Statistical analyses were computed using the GraphPad Prism software version 5 (GraphPad Software Inc., La Jolla, CA). A P value <.05 was considered significant.
Results
Purified hnDNA or individual human histone proteins, but not intact NETs, trigger coagulation
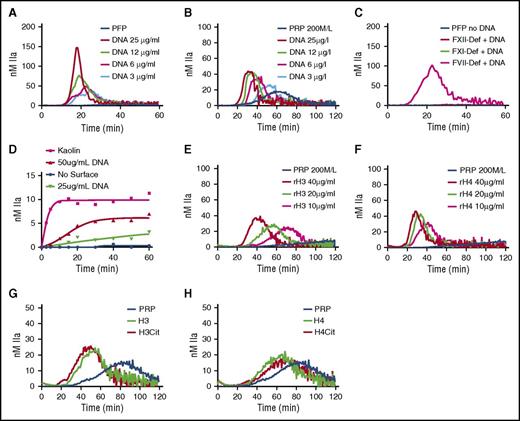
TG was monitored in recalcified PFP or PRP in the presence of increasing concentration of hnDNA. No TG was observed in the baseline PFP (Figure 2A) or PRP (Figure 2B). However, a concentration-dependent increase in TG was observed in the presence of hnDNA in PFP (Figure 2A) and in PRP, although with different kinetics (Figure 2B). In the presence of 30 µg/mL hnDNA, TG was observed in FVII deficient, but not in FXII deficient or FXI deficient (Figure 2C), indicating that the triggering effect of hnDNA on coagulation is mediated via contact activation. No TG was observed in either deficient plasma in the absence of hnDNA (Figure 2C). hnDNA also showed a dose-dependent activation of the contact pathway in a purified system composed of contact system proteins (Figure 2D).
hnDNA and individual human histone H3 and H4 trigger coagulation in plasma. TG in recalcified normal PFP (A) and normal PRP (B) containing hnDNA. TG in recalcified FXII-deficient (FXII-Def), FXI-deficient (FXI-Def), or FVII-deficient (FVII-Def) PFP in the presence or absence of 30 µg/mL of hnDNA (C). No TG was observed in any individual deficient plasma after recalcification in the absence of DNA, represented by a single flat curve (PFP no DNA, panel C). Quantification of FXIa-AT after activation of the contact system by hnDNA in the synthetic contact system activation assay as described in “Methods” (D). TG in recalcified PRP containing recombinant human histone H3 (rH3) (E) or recombinant human histone H4 (rH4) (F). Effect of citrullination of histones H3 (G) and H4 (H) on TG in recalcified PRP. All the figures are representative of at least 3 independent experiments.
hnDNA and individual human histone H3 and H4 trigger coagulation in plasma. TG in recalcified normal PFP (A) and normal PRP (B) containing hnDNA. TG in recalcified FXII-deficient (FXII-Def), FXI-deficient (FXI-Def), or FVII-deficient (FVII-Def) PFP in the presence or absence of 30 µg/mL of hnDNA (C). No TG was observed in any individual deficient plasma after recalcification in the absence of DNA, represented by a single flat curve (PFP no DNA, panel C). Quantification of FXIa-AT after activation of the contact system by hnDNA in the synthetic contact system activation assay as described in “Methods” (D). TG in recalcified PRP containing recombinant human histone H3 (rH3) (E) or recombinant human histone H4 (rH4) (F). Effect of citrullination of histones H3 (G) and H4 (H) on TG in recalcified PRP. All the figures are representative of at least 3 independent experiments.
Histones are major components of NETs. Recombinant human histones H3 or H4 dose-dependently triggered TG in recalcified PRP (Figure 2E-F, respectively), but not in recalcified PFP, indicating the dependency on platelets, as previously described.7 Citrullination of recombinant human histones H3 and H4 by incubation with PAD4 enzyme in the presence of calcium as previously reported22 did not modify their respective procoagulant activities in PFP or PRP (Figure 2G-H).
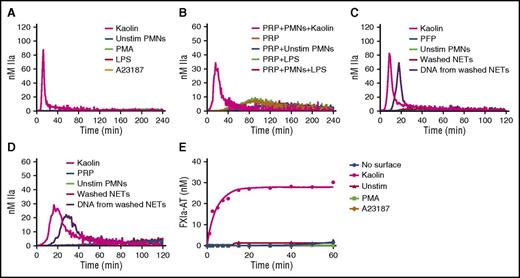
To assess the effect of intact NETs, TG was monitored in recalcified PFP containing NETs released from stimulated human neutrophils. Two models of NETosis, which differ by the inducing stimuli, the mechanisms and lag time for NET release, and the fate of neutrophils, have been proposed.23 Suicidal NETosis, whereby neutrophils die after NET release, is reproduced by PMA stimulation, whereas vital NETosis is reproduced by more biological inducers such as lipopolysaccharide.24 Because PMA- and ionophore-stimulated platelets can also trigger TG in plasma (supplemental Data 1, available on the Blood Web site), platelet contamination had to be avoided during neutrophil isolation. Indeed, neutrophil isolates contained undetectable platelet or red blood cell contamination (supplemental Data 2). In addition, washed NETs were used in PRP to avoid PMA- or ionophore-induced TG mediated by platelet activation. The measured levels of cell-free DNA and nucleosomes showed that the final preparation contained abundant NETs (supplemental Data 3). In contrast to kaolin in PFP or in PRP, no thrombin was formed in the presence of intact NETs generated in situ after incubation for 4 hours regardless of the NETs inducer (Figure 3A-B). Similarly, no TG was observed when washed NETs were incubated for 2 hours with PFP (Figure 3C) or PRP (Figure 3D) (data shown for neutrophil count = 10 000/µL). Interestingly, DNA purified from the same amount of washed NETs triggered TG in PFP and PRP (Figure 3C-D). Treatment of intact NETs with 20 µg/mL DNAse for 3 hours at 37°C did not trigger TG in PFP or PRP, and the level of DNA isolated from DNAse-treated NETs was markedly reduced (data not shown). Consistently, intact NETs did not activate the contact system in the SCSA (Figure 3E).
Human intact NETs do not trigger coagulation in plasma or in a purified contact system. (A) TG in recalcified PFP containing unstimulated neutrophils (Unstim PMNs) or NETs prepared by stimulation of normal human neutrophils with PMA (600 nM), ionophore (A23187; 5 µM), or LPS (5 µg/mL) (B). TG was performed in recalcified plasma containing LPS-induced NETs in the presence of platelets (200 000/µL). TG was performed in PFP (C) and PRP (D) unstimulated neutrophils, washed NETs, or DNA purified from the same amount of washed NETs. NETs were washed to remove PMA or ionophore before being added to plasma (see “Methods”). Quantification of FXIa-AT in the SCSA in the presence of NETs (E). All figures are representative of at least 3 independent experiments. LPS, lipopolysaccharide.
Human intact NETs do not trigger coagulation in plasma or in a purified contact system. (A) TG in recalcified PFP containing unstimulated neutrophils (Unstim PMNs) or NETs prepared by stimulation of normal human neutrophils with PMA (600 nM), ionophore (A23187; 5 µM), or LPS (5 µg/mL) (B). TG was performed in recalcified plasma containing LPS-induced NETs in the presence of platelets (200 000/µL). TG was performed in PFP (C) and PRP (D) unstimulated neutrophils, washed NETs, or DNA purified from the same amount of washed NETs. NETs were washed to remove PMA or ionophore before being added to plasma (see “Methods”). Quantification of FXIa-AT in the SCSA in the presence of NETs (E). All figures are representative of at least 3 independent experiments. LPS, lipopolysaccharide.
Purified hnDNA, but not intact NETs, amplifies low-TF-initiated TG in PFP
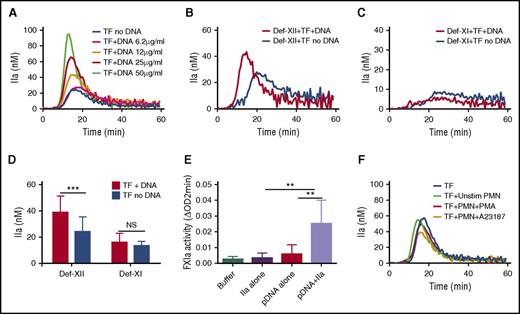
We next tested the ability of hnDNA or intact NETs to amplify low-TF-initiated TG in recalcified human normal PFP. In these experiments, whole blood was collected in the presence of 0.1 mg/mL CTI. A dose-dependent amplification of TG was observed in the presence of hnDNA (Figure 4A). The procoagulant effect of hnDNA was still observed in FXII-deficient plasma (Figure 4B,D). Specifically, the mean peak TG was increased by 60% in the presence of 25 µg/mL hnDNA in FXII-deficient PFP (38.9 ± 12.3 vs 24.4 ± 11.0, respectively; P < .001), whereas no difference was observed in TG in FXI-deficient PFP (16.2 ± 6.7 vs 13.4 ± 3.4, respectively; P = 0.279) (Figure 4C-D). These results suggest that the amplification effect of hnDNA on low-TF-initiated TG in PFP is dependent on FXI, but not on FXII. To further confirm the possibility that hnDNA enhances FXIa generation by thrombin, the chromogenic thrombin-dependent FXIa generation assay was performed in the presence or absence of 50 µg/mL hnDNA. The results showed that hnDNA amplified thrombin-dependent FXIa by 8- to 12-fold compared with thrombin alone (Figure 4E). However, when low-TF-initiated TG was monitored in normal PFP in the presence or absence of intact NETs, no amplification effect was observed, regardless of the number of neutrophils or their stimulation by PMA or ionophore (Figure 4F; data shown only for neutrophil counts = 12 000/µL).
Purified hnDNA enhances low-concentration TF–initiated TG by amplifying thrombin-dependent FXI activation. Amplification effect of hnDNA on TG triggered by 1 pM TF in PFP (A) or in FXII-deficient plasma (Def-XII; 25 μg/mL hnDNA) (B) or in FXI-deficient plasma (Def-XI; 25 μg/mL hnDNA) (C). (D) Comparison of peak TG (mean + standard deviation) in the presence or absence of 25 μg/mL hnDNA in FXII- or FXI-deficient plasma. (E) Amplification effect of 50 μg/mL hnDNA on FXIa generation following 10 nM α-thrombin (IIa)–dependent activation of 30 nM FXI in the presence of physiologic concentration of HMWK in buffer. (F) Absence of an amplification effect of intact NETs on 1 pM TF-initiated TG in PFP. A23187, ionophore (5 μM); PMN, human neutrophils. PMA (600 nM). Panels A-C and F are representative of independent experiments performed at least 3 times. Mean values for 3 independent experiments in panels C and D.
Purified hnDNA enhances low-concentration TF–initiated TG by amplifying thrombin-dependent FXI activation. Amplification effect of hnDNA on TG triggered by 1 pM TF in PFP (A) or in FXII-deficient plasma (Def-XII; 25 μg/mL hnDNA) (B) or in FXI-deficient plasma (Def-XI; 25 μg/mL hnDNA) (C). (D) Comparison of peak TG (mean + standard deviation) in the presence or absence of 25 μg/mL hnDNA in FXII- or FXI-deficient plasma. (E) Amplification effect of 50 μg/mL hnDNA on FXIa generation following 10 nM α-thrombin (IIa)–dependent activation of 30 nM FXI in the presence of physiologic concentration of HMWK in buffer. (F) Absence of an amplification effect of intact NETs on 1 pM TF-initiated TG in PFP. A23187, ionophore (5 μM); PMN, human neutrophils. PMA (600 nM). Panels A-C and F are representative of independent experiments performed at least 3 times. Mean values for 3 independent experiments in panels C and D.
Assembly of core histones abrogates individual histone protein–mediated activation of coagulation
Individual histone proteins assemble in heterodimers of H2A-H2B or H3-H4. These heterodimers further assemble in tetramers and ultimately octamers to form the core histones25 that are found in nucleosomes and chromatin. TG was performed in recalcified PRP or PFP in the presence of recombinant human core histones or in the presence of a mixture of natural histones purified from calf thymus. In contrast to individual histone proteins H3 and H4, both artificial and natural core histones failed to trigger TG in PRP (Figure 5A-B, respectively). No TG was observed in recalcified PFP containing either type of core histones (data not shown). These results indicate that individual histone proteins H3 and H4 lose their procoagulant activity when they assemble to form the core histone structure.
Individual human histones H3 and H4, but not octameric core histones, trigger TG in PRP. TG performed in recalcified PRP containing individual human histone proteins was compared with that of recombinant core histone octamers (A) or purified calf thymus histones (B). CThist, mixture of purified calf thymus histones; rOctamer, octameric core histone reconstituted with recombinant human histone proteins. Panels A and B are representative of 3 independent experiments.
Individual human histones H3 and H4, but not octameric core histones, trigger TG in PRP. TG performed in recalcified PRP containing individual human histone proteins was compared with that of recombinant core histone octamers (A) or purified calf thymus histones (B). CThist, mixture of purified calf thymus histones; rOctamer, octameric core histone reconstituted with recombinant human histone proteins. Panels A and B are representative of 3 independent experiments.
Assembly of single nucleosomes abrogates the procoagulant effects of purified DNA
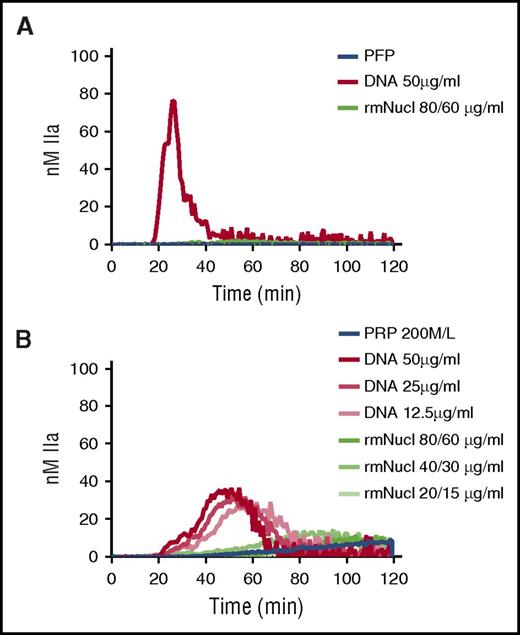
A single nucleosome particle (“mononucleosome”) consists of an octameric core of histones around which 147 bp of DNA are wrapped in 1.65 turns.25 Mononucleosomes can be reconstructed in vitro using recombinant histones and tandemly repeated positioning DNA.26 TG was performed in recalcified PFP or PRP in the presence of hnDNA or mononucleosomes reconstructed in vitro using recombinant human histones. Purified hnDNA triggered TG both in PFP and PRP as expected, whereas no significant thrombin was generated in either plasma type in the presence of mononucleosomes containing concentrations of histones and DNA known to activate coagulation (Figure 6A-B).
Purified hnDNA, but not mononucleosome particles, trigger TG in plasma. (A) TG generation performed in recalcified PFP containing hnDNA or recombinant mononucleosomes. (B) TG generation performed in recalcified PRP containing hnDNA or recombinant mononucleosomes. rmNucl, mononucleosomes consisting of reconstituted recombinant human histone proteins and DNA. The unit of rmNucl is expressed as the ratio of final concentrations of histones to DNA. Panels A and B are representative of 3 independent experiments.
Purified hnDNA, but not mononucleosome particles, trigger TG in plasma. (A) TG generation performed in recalcified PFP containing hnDNA or recombinant mononucleosomes. (B) TG generation performed in recalcified PRP containing hnDNA or recombinant mononucleosomes. rmNucl, mononucleosomes consisting of reconstituted recombinant human histone proteins and DNA. The unit of rmNucl is expressed as the ratio of final concentrations of histones to DNA. Panels A and B are representative of 3 independent experiments.
Chromatin reconstitution abolishes the procoagulant effects of purified hnDNA
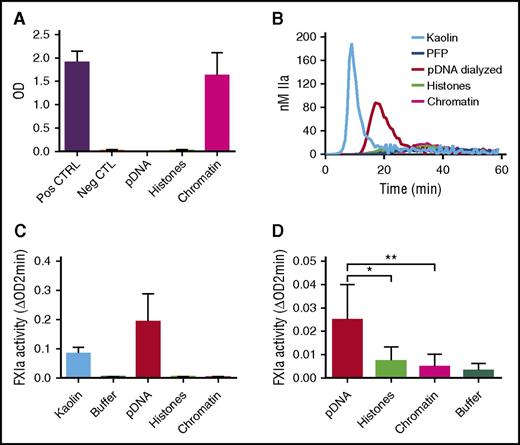
We next reconstituted chromatin with genomic DNA and histones that were separately isolated from normal human neutrophils (see “Methods”). Nucleosomes were detected in samples containing reconstituted chromatin and not in those containing hnDNA alone or histones alone, confirming successful chromatin reconstitution (Figure 7A). In contrast to hnDNA, reconstituted chromatin did not migrate during agarose gel electrophoresis, indicating the neutralization of hnDNA negative charge (supplemental Data 4). DNA was quantified in the dialyzed samples to ensure it was fully recovered, as was indeed the case (data not shown). In contrast to dialyzed hnDNA, reconstituted chromatin did not trigger TG in recalcified PFP, nor did it activate the contact system or amplify thrombin-dependent FXIa generation in buffer (Figure 7B-D, respectively). These results demonstrate that in vitro reconstitution of stable DNA-histone complexes using isolated hnDNA and histones to mimic natural chromatin abolishes the enhancing effects of hnDNA on coagulation.
Effect of chromatin reconstitution on purified DNA-dependent activation of coagulation. Chromatin was reconstituted using DNA and histones purified from normal human neutrophils. (A) Detection of nucleosomes in samples following reconstitution; positive and negative controls are provided with the ELISA kit. (B) TG performed in recalcified PFP in the presence or absence of 30 μg/mL kaolin (positive control), 50 μg/mL dialyzed hnDNA, dialyzed histones, or reconstituted chromatin. (C) Effect of dialyzed hnDNA, histones, and reconstituted chromatin on FXIa generation in the presence of physiologic concentrations of FXII, FXI, and HMWK in buffer. (D) Effect of dialyzed hnDNA, histones, and reconstituted chromatin on 10 nM α-thrombin-dependent activation of FXI (30 nM) in buffer in the presence of physiologic concentrations of HMWK. hnDNA, purified hnDNA. Panel B is representative of 3 independent experiments. Mean ± standard deviation of 3 independent experiments in panels A, C, and D.
Effect of chromatin reconstitution on purified DNA-dependent activation of coagulation. Chromatin was reconstituted using DNA and histones purified from normal human neutrophils. (A) Detection of nucleosomes in samples following reconstitution; positive and negative controls are provided with the ELISA kit. (B) TG performed in recalcified PFP in the presence or absence of 30 μg/mL kaolin (positive control), 50 μg/mL dialyzed hnDNA, dialyzed histones, or reconstituted chromatin. (C) Effect of dialyzed hnDNA, histones, and reconstituted chromatin on FXIa generation in the presence of physiologic concentrations of FXII, FXI, and HMWK in buffer. (D) Effect of dialyzed hnDNA, histones, and reconstituted chromatin on 10 nM α-thrombin-dependent activation of FXI (30 nM) in buffer in the presence of physiologic concentrations of HMWK. hnDNA, purified hnDNA. Panel B is representative of 3 independent experiments. Mean ± standard deviation of 3 independent experiments in panels A, C, and D.
Discussion
This study includes a comprehensive assessment of the effect of human NETs generated in vitro and their components (at different levels of assembly) on coagulation in human plasma and purified systems. Using a robust and reproducible method to generate NETs from normal human neutrophils, the data provide evidence that in contrast to the procoagulant effect of recombinant human histone proteins H3 and H4 in PRP and hnDNA in both PFP and PRP, intact NETs failed to initiate the contact pathway and failed to amplify both thrombin-dependent FXIa generation and low-TF-initiated TG in plasma. Interestingly, chromatin reconstituted from neutrophil histones and hnDNA, and its structural units (nucleosomes), abolished the enhancing effect of hnDNA on coagulation and displayed similar patterns as intact NETs. Likewise, assembly of individual human histone proteins to form the octameric histone core abrogated the procoagulant effect of histone proteins H3 and H4 in PRP.
High concentrations of purified DNA from various cellular sources have been shown to trigger the intrinsic pathway of coagulation in PPP and in purified systems.10-12,27 This study adds that the procoagulant effect of hnDNA can be seen at DNA concentrations as low as 3 µg/mL in plasma. In addition to confirming the known effects of purified hnDNA on FXII-dependent contact activation, we demonstrate for the first time that hnDNA can also amplify TF-initiated TG in plasma. Our data suggest that this effect is dependent on FXI, and not FXII. However, because CTI is an incomplete inhibitor of FXIIa, we cannot fully exclude the contribution of FXIIa in the amplification effect observed in normal PFP. Consistent with our results, Vu et al recently reported an amplification effect of DNA purified from A549 cells on FXIIa- and thrombin-dependent FXIa generation in buffer, to a magnitude close to what was found in this study.10 Furthermore, they demonstrated that both FXI and thrombin bind purified DNA, which was therefore proposed as a cofactor for the amplification of thrombin-dependent FXIa generation.10 The molecular interactions involved in this effect are likely charge dependent because other polyanionic compounds, including dextran sulfate, sulfatides, glycosaminoglycans, and polyphosphates, can similarly amplify FXI activation by thrombin in buffer or in plasma.10,28-30 In contrast, intact NETs did not amplify low-TF-initiated TG in plasma, and the amplifying effect of hnDNA in buffer was abolished when it was used to reconstitute chromatin. Indeed, reconstitution of chromatin affected the electrophoretic mobility of hnDNA (supplemental Data 4) likely by reducing its net negative charge as a result of interactions with positively charged histones. However, the ability of intact NETs or reconstituted chromatin to bind thrombin or FXI was not tested in this study. Inflammation and TF upregulation are hallmarks of clinical conditions in which markers of in vivo NET release have also been reported, including sepsis, cancer, autoimmune diseases, and sickle-cell disease.31-33 It could be speculated that both intrinsic and extrinsic pathways contribute synergistically to enhance coagulation in these conditions, where TF may be the primary trigger and free DNA the cofactor that amplifies FXIa generation. Therefore, blocking FXIa or its generation presents a potential target to minimize the risk of thrombosis.
Consistent with other studies using different sources of histones,7 we found that individual human histone H3 and H4 triggered TG in PRP. This effect has been attributed to histone-mediated platelet activation through the binding of histones to platelet Toll-like receptors 2 and 47,14 or through the binding to membrane phosphatidylserine,34 possibly inducing changes in the membrane conductance of ions.35 However, the assembly of individual histone proteins to form the core octameric structure apparently interferes with histone-induced platelet activation as shown by the absence of TG in PRP in the presence of the octameric core histones. Likewise, reconstituted chromatin did not reproduce the procoagulant effect of hnDNA or histones. Charge neutralization is likely the reason why intact NETs, which are built on extracellular chromatin, do not expose sufficient net negative charge to activate the contact system. The binding of histone proteins to one another and to DNA may thus represent a natural protective mechanism against the detrimental effect of these individual components in their free state.
The absence of coagulation activation by intact NETs in this study is unlikely to be explained by an insufficient quantity of NETs, because hnDNA extracted from the same amount of NETs did activate coagulation. It is important to note that although this study addresses the direct effect of human NETs on coagulation and brings greater clarity to current knowledge in this area, the data are generated exclusively from in vitro studies. It has previously been reported that NETs promote thrombosis in vivo using some animal models,2-4 although the contribution of neutrophils and NETs could not be demonstrated in another mouse model of spontaneous venous thrombosis.36 The in vitro systems used in this study do not account for the participation of other potentially relevant cells or the effects of flow in vivo, where intravascular NETs may disrupt laminar flow, injure the endothelium, and/or recruit platelets, red blood cells, or TF-containing microparticles. Any or all of these factors could potentially contribute to thrombosis.2,3,24,37 Furthermore, breakdown of NETs by endonucleases and proteases could lead to the release of individual NET components (DNA or histones) that may activate coagulation. For example, plasmin has been shown to degrade chromatin-bound histones in vitro, potentially exposing histone-free DNA.38 Finally, and perhaps most importantly, inferring knowledge from in vitro studies using human NET components to any in vivo effects of NETs (or NET components) in animal models can be misleading. Further studies are therefore required to address possible interspecies differences in NET and NET component-induced procoagulant activities.
In summary, although NETs are present in the developing thrombus, and their degradation by DNAase prevents thrombosis in animal models, human intact NETs do not directly trigger coagulation in vitro. This is relevant because some mechanisms that prevent NET formation or dismantle the NET scaffold could be an effective antithrombotic strategy, whereas those that interfere with downstream NET-induced TG might not. Further in vitro studies are needed to elucidate how intact NETs interact with coagulation in a more complete system that also includes other blood cells, vascular endothelium, and flow.
Presented in abstract form at the 57th annual meeting of the American Society of Hematology, Orlando, FL, 7 December 2015.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Michael Henderson, Leila ElSherif, Anton Ilich, and Shaobin Wang for their technical assistance and advice.
This work was supported by grants from the National Heart, Lung, and Blood Institute, National Institutes of Health (T32HL007149 and UO1HL117659) and by an American Heart Association fellowship (16POST30230002) (E.S.).
Authorship
Contribution: D.F.N., M.F.W., N.S.K., R.P., and D.M.M. designed the study; D.F.N., Y.-B.Y., E.S., and M.F.W. performed the experiments; D.F.N. analyzed the data and wrote the manuscript; and M.F.W., N.S.K., D.M.M., R.P., and Y.-B.Y. edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nigel S. Key, 1079 Genetic Medicine Building, CB #7035, 120 Mason Farm Rd, Chapel Hill, NC 27599; e-mail: nigel_key@med.unc.edu.