In this issue of Blood, Mar et al describe the effect of inactivating mutations of the histone methyltransferase (HMT) SETD2 in accelerating leukemia pathogenesis and conferring therapy resistance.1 Mutations of epigenetic regulators are among the commonest lesions in malignancy. These include mutations of HMTs that modify histone tails protruding from the nucleosome. For example, inactivation of EZH2, responsible for histone H3 lysine 27 trimethylation (H3K27me3), a modification associated with gene silencing, occurs in myelodysplasia and acute myeloid leukemia (AML), whereas mutation of KMT2D, responsible for the H3K4me1 modification found at enhancers, is common in lymphoma. These enzymes change the chemical composition of chromatin at gene regulatory sites, affecting transcriptional initiation. By contrast, SETD2 has a role downstream of the transcriptional start site (TSS).
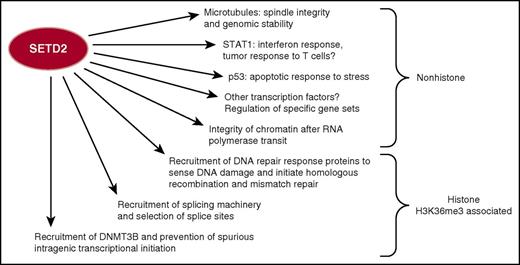
SETD2, the sole known HMT to create the histone H3 lysine 36 trimethyl (H3K36me3) modification through this histone change, mediates multiple molecular processes of gene regulation and DNA damage response. SETD2 also has nonhistone substrates with important roles in cellular homeostasis. Loss of SETD2 activity therefore may interrupt a host of critical processes that contribute to malignancy.
SETD2, the sole known HMT to create the histone H3 lysine 36 trimethyl (H3K36me3) modification through this histone change, mediates multiple molecular processes of gene regulation and DNA damage response. SETD2 also has nonhistone substrates with important roles in cellular homeostasis. Loss of SETD2 activity therefore may interrupt a host of critical processes that contribute to malignancy.
Mutations of SETD2, including truncating mutations, are commonest in clear cell renal carcinoma (∼20%) and are found in 5% to 10% of cases of a wide variety of tumors, including melanoma, bladder, lung, and uterine (www.cbioportal.org). SETD2 mutations were found in 12% of cases of B-cell acute lymphoblastic leukemia, 1% to 2% of cases of B-cell lymphoma, chronic lymphocytic leukemia, and AML, and occasionally cases of myeloproliferative neoplasm. SETD2 is the sole mammalian HMT that catalyzes H3K36 trimethylation (H3K36me3). SETD2 associates with elongating RNA polymerase, creating H3K36me3-modified nucleosomes 3′ to the TSS that serve as docking sites for the FACT histone chaperone and assembly complex. Thus, H3K36me3 methylation by SETD2 and recruitment of tight arrays of nucleosomes prevent the spurious reinitiation of transcription within gene bodies. H3K36me3 also recruits DNMT3B, leading to the dense methylation of gene bodies, which reinforces intragenic silencing.2 In addition, H3K36me3 helps direct the splicing machinery to intron/exon boundaries.
SETD2 has an important role in DNA repair. The MSH6 protein recognizes H3K36me3, and SETD2 activity is required for base-mismatch repair.3 SETD2 and H3K36me3 are also required for recruitment of RAD51 and LEDGF to double-stranded DNA breaks, promoting repair of active genes by homologous recombination.4 Furthermore, SETD2 depletion/mutation leads to decreased recruitment of DNA replication machinery and replication fork instability. SETD2 also functions outside of the nucleus, methylating tubulin, and loss of SETD2 leads to mitotic abnormalities.5 SETD2 also methylates STAT1 and is important for interferon-mediated antiviral6 and potentially immune surveillance response. In addition, SETD2 can bind and alter p53 activity.
The many functions of SETD2 suggest how its inactivation might contribute to malignancy (see figure). In renal cancer, loss of SETD2 led to depletion of nucleosomes, loss of DNA methylation, aberrant splicing, and abnormal intragenic RNAs. SETD2 mutant renal cancer cell lines are also deficient in DNA repair.7 Previous studies in blood malignancy showed that knockdown of Setd2 in an MLL-fusion AML led to increased tumor cell growth. This was associated with increased expression of mTOR and STAT pathways.8 SETD2 mutations were found in 25% of the rare hepatosplenic γδ T-cell lymphoma (HSTL), and knockdown of SETD2 in HSTL cell lines increased cell growth and altered expression of cell-cycle genes. SETD2 mutation is also found in enteropathy-associated T-cell lymphoma. The predilection of the mutation for these tumors may be explained by the observation from knockout models that SETD2 controls the ratio of γδ vs αβ T cells.9 However, the mechanism by which the depletion of H3K36me3 alters a specific gene and developmental program remains unexplained.
The work of Mar et al further implicates SETD2 as a tumor suppressor. Using CRISPR/CAS9 to generate AML cells with disruption of SETD2, they also observed that DNA damage response and repair were compromised. In a murine model in which bone marrow was transformed with MLL-AF9, disruption of both Setd2 alleles inhibited leukemogenesis, potentially because of excessive DNA damage and impaired replication. This effect may be cell type specific because renal cancer often suffers complete loss of SETD2, whereas mutations in leukemia are heterozygous. Loss of one allele of Setd2 led to decreased leukemia latency and resistance to chemotherapy. How loss of a single allele accelerates disease remains to be determined but may be due to deregulation of subsets of growth-activating genes or increased DNA damage and accumulation of secondary mutations.
The epigenetic lesions in malignancy are vexing because targeted therapy cannot be applied to the common loss-of-function mutations. In the case of the HMTs, rebalancing chromatin patterns by targeting opposing enzymes can be considered. For example, inactivation of the H3K27me3 histone demethylase KDM6A in bladder cancer or myeloma can be approached by inhibition of EZH2. Consequently, Mar et al applied an inhibitor of the H3K36 demethylase KDM4A to SETD2 mutant cells and observed a rise in H3K36me3 levels. How this occurred in a cell with compound heterozygous mutation of SETD2 is unclear and may be a consequence of residual activity of SETD2 or compensation by an as yet unidentified HMT. The elevated H3K36me3 levels tracked with an increase in sensitivity to chemotherapy in vitro, and the combination of demethylase inhibitor and chemotherapy could acutely kill more leukemia cells in vivo. However, this was not sufficient to extend animal life span, perhaps because of insufficient pharmacodynamics or pharmacokinetics, the ability of the inhibitor to affect other demethylases, or other aspects of AML biology affected by Setd2 loss.
SETD2 mutations are associated with poor prognosis in malignancy, and although rebalancing chromatin function is a rational approach, it may not be the answer. In SETD2 mutant renal cancer, ribonucleotide reductase 2 (RRM2) levels were low and further decreased by a Wee1 inhibitor, representing a synthetic lethal approach.10 However, in the AML model, RRM2 levels were unchanged, again pointing to potentially indirect and cell type–specific effects of SETD2 loss on gene expression conceivably through nonchromatin mechanisms, such as modification of transcription factor activity. With a genetically faithful model of SETD2-associated malignancy in hand, further screens for vulnerabilities using gene editing are warranted. Thorough analysis of these mice for changes in gene expression and chromatin modifications, p53 target expression, and interferon pathways may lead to further clues to the aggressive biology generated by SETD2 loss.
Conflict-of-interest disclosure: The author declares no competing financial interests.