Abstract
Background: Acute myeloid leukemia (AML) is the cancer of the myeloid precursor stem cells. AML was historically treated with combinations of cytarabine (ara-C) and anthracyclines. Previous studies showed that polymorphism in genes known to be involved in cytarabine pathway can affect patient's drug response and may explains part of the inter-patient variability in treatment response.
Patients and methods: AML patients (n= 89) treated under St. Jude AML97 clinical trial were included in the study. Intracellular levels of ara-CTP the active metabolite of ara-C was quantitated in leukemic cells obtained 24hrs after initiation of cytarabine infusion. Morphological response to induction 1 therapy was assessed in bone marrow aspirate obtained on day 15. Genomic DNA was tested for SNPs in DCK and CMPK1 required for the drug activation, CDA, DCTD, NT5C2, NT5C3A, RRM1 and RRM2 involved in Cytarabine inactivation and the drug transporters SLC29A1, SLC28A1 and SLC28A3. Additionally, CTPS1, NME1-NME2 and NME3 that are important for the synthesis of nucleoside triphosphates other than ATP were also included. Overall 104 SNPs in genes indicated above were genotyped using Sequenom (CA, USA) based genotyping that uses MALDI-TOF-based chemistry at University of Minnesota, Biomedical Genomics Center.
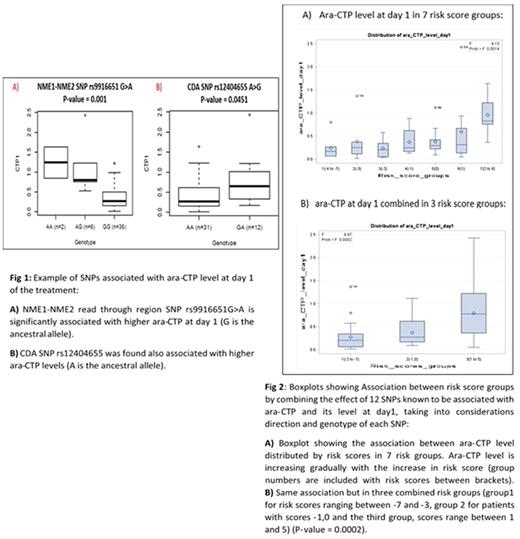
Results: Tweleve SNPs demonstrated a statistically significant association with ara-CTP level at day 1 post treatment (P<0.05). We found SNP rs4643786 in DCK gene significantly associated with lower ara-CTP levels and poor outcomes in white patients. CDA SNP rs12404655, was associated with high ara-CTP level at day 1 and better treatment outcomes (Fig 1B). CMPK1 SNP rs7543016, a missense cleavage site variant was associated with lower ara-CTP levels. In addition, SNP rs9916651 in NME1-NME2 was predictive of higher ara-CTP levels (Fig 1A).
We observed 7 SNPs to be significantly associated with the treatment response after induction 1 this included a previously reported NT5C2 SNP rs11598702 and 4 SNPs in DCTD that were associated with poor response and occured in linkage disequilibrium with each other's (r2=1: rs3190314, rs1130902, rs7277 and rs851).
Using the genotypes of SNPs associated with intracellular ara-CTP levels we created a risk score by combining the effect of SNPs reported in each patient. For this we took into consideration the direction of genotype association for each SNP. For example, if the SNP is associated with higher ara-CTP level, it takes a score ranging between 0 and +2 (0 if the patient has normal alleles, 1 if he/she is heterozygous and 2 if homozygous for the SNP). If the SNP is associated with lower ara-CTP level, it takes a score ranging between 0 and -2. Risk scores ranging between -7 and 5 were obtained after this analysis. We further compressed the risk groups into three categories (risk score ≥1, risk score 0, -1 and risk score ≤ -2 (Figure 2). Increased value of the risk score was associated with increased intracellular ara-CTP level suggesting a cumulative or synergistic effect of the reported SNPs.
Conclusion: Our results suggest that SNPs in genes related to cytarabine pathway can significantly affect level of ara-CTP inside the leukemic cells. To comprehensively include the potentially significant SNPs we developed a risk score for each patient by taking into account the directional genotype for each SNP in each patient. We hope that this score although require validation in other cohorts will help in identification of patients with potentially lower levels of active form of ara-CTP in the leukemic cells. We would also like to point out that it is very challenging to perform this study as it requires obtaining bone marrow samples for determining intracellular levels of ara-CTP after day 1 of initiation of chemotherapy, which is not a standard procedure. To the best of our knowledge this is the only clinical cohort of pediatric AML patients where intracellular ara-CTP levels post ara-C treatment are available and we leveraged this to evaluate genetic variants. Our result would provide significant information in further understanding the pharmacogenomics of cytarabine in AML.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.