Abstract
Background
Venous thromboembolism (VTE) in cancer significantly contributes to morbidity and a worse overall prognosis. A validated risk tool for general oncology patients exists but novel biomarkers with greater sensitivity and specificity are urgently needed. We embarked on a comprehensive RNA sequencing strategy, focusing on a single solid tumor (lung cancer) to identify novel biomarkers of cancer-associated VTE.
Methods
We identified 75 lung cancer patients at the Cleveland Clinic who underwent surgical excision or biopsy of primary lung mass and had tissue collected on an IRB-approved protocol. Of these, we categorized patients into those who developed subsequent VTE (N=15) and those who did not (N=60). Nine patients with VTE were then propensity-matched (score difference of <5%) with nine VTE-negative patients based on age, gender, race, history of prior cancer, date of cancer diagnosis, stage, histology, number of lines of chemotherapy, and length of follow up.
RNA was extracted, prepared into mRNA libraries, and sequenced by Illumina HiSeq resulting in paired 100nt reads, and subjected to quality control. RNA reads were aligned to hg19 genome assembly using Mapsplice. Gene expression was quantified for transcript models corresponding to TCGA GAF2.13, using RSEM4 and normalized within-sample to a fixed upper quartile. For gene level analyses, expression values of zero were set to overall minimum value, and all data were log2 transformed. The log2 fold-change and adjusted p-values (using Benjamini-Hochberg procedure) were calculated using linear models in combination with moderated t-statistic. Finally, association between single-sample gene set enrichment analysis (GSEA) profiles for each gene set and binary categorical variables were determined using an information-based similarity metric (RNMI).
Results
The final study population included twelve patients with lung tissue RNA samples meeting criteria for quality control (six each with or without VTE) with a median age of 67 years. 58% of patients had metastatic disease, 8% stage I or II, and 25% had stage III disease. Histology included 58% adenocarcinoma, 25% squamous, and 16% mixed/other. 67% of all patients received carboplatin doublet therapy as first line treatment, 25% received no chemotherapy, and 8% received immunotherapy. Of the six patients with VTE, 50% had pulmonary embolism, 33% had lower extremity deep venous thromboses, and 17% had catheter-associated thrombus.
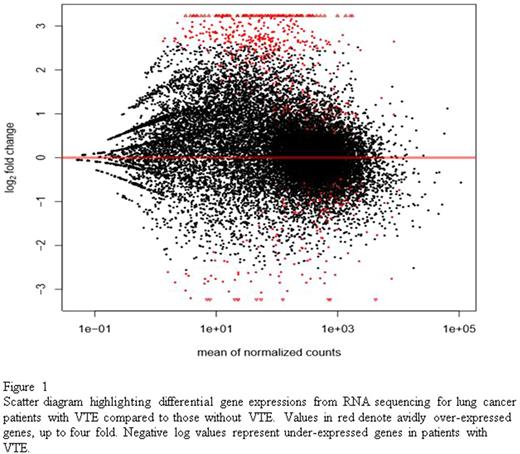
RNA sequencing revealed a total of 1037 genes with statistically significant (q <0.05) differential expression. In patients with VTE, 869 genes were over-expressed and 168 were under-expressed compared to patients without VTE. Figure 1 shows a scatter diagram reflecting gene expression values between the two cohorts. Genes of interest over-expressed in patients with VTE included complement receptors: CECR1 (4.9 fold elevation, q = 0.000008) and CR1 (4.2 fold elevation, q = 0.0004), myocardial infarction transcript MIAT (4.8 fold elevation, q = 0.000003), RAS activating pathways SHC4 (4.6 fold elevation, q = 0.00003), cytokine receptors responsible for activating and regulating inflammatory response: NLRP14 (3.8 fold elevation, q = 0.0001), IL5RA (3.6 fold elevation, q = 0.006), IL16 (3.4 fold elevation, q = 0.002), CLNK (3.6 fold elevation, q = 0.004), and XCL1 (3.5 fold elevation, q = 0.008). In lung cancer patients with VTE, GSEA revealed over-expression of genes involved in IL2-STAT5 signaling, IL6-JAK-STAT3 signaling, IFN-gamma response, inflammatory response, KRAS signaling, and complement pathway.
In lung cancer patients without VTE, GSEA revealed over-expression of genes involved in apoptosis and epithelial mesenchymal transition pathways.
Conclusions
We have identified a novel gene expression profile in lung cancer patients who have experienced VTE. Notably, complement and inflammatory pathways are upregulated in lung cancers that develop VTE. These differentially expressed genes and pathways provide novel biologic insight into cancer-associated VTE and may lead to development of new risk stratification as well as therapeutic strategies.
Khorana: Leo: Consultancy, Honoraria, Research Funding; Bayer: Consultancy, Honoraria; Sanofi: Consultancy, Honoraria; Halozyme: Consultancy, Honoraria; Roche: Consultancy, Honoraria; Pfizer: Consultancy, Honoraria; Janssen Scientific Affairs, LLC: Consultancy, Honoraria, Research Funding; Amgen: Consultancy, Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.