Abstract
Over the last 10 years, DNA sequencing has revolutionized our understanding of the pathogenesis of this disease, establishing that MDS arises through the sequential acquisition of somatic mutations in a set of recurrently involved genes. It is now known that spliceosome genes represent the most common targets of mutations in patients with MDS. U2AF1 is a common (about 10%) mutated spliceosome gene in MDS. Understanding the clinical features and biological implications of U2AF1 mutation (U2AF1 MT) in MDS is only just beginning.
To investigate the clinical impact of U2AF1 MT in MDS, we studied a cohort of 511 patients who were performed 112-targeted gene sequencing. We compared the mutational landscapes of U2AF1 MT subjects, their clinical associations, survival, and clonal hierarchies. Our aims were to provide insights into U2AF1 MT MDS pathogenesis, especially with respect to the clonal architecture and different mutation type of U2AF1 MT subjects, and to determine whether U2AF1 MT should be grouped or considered separately.
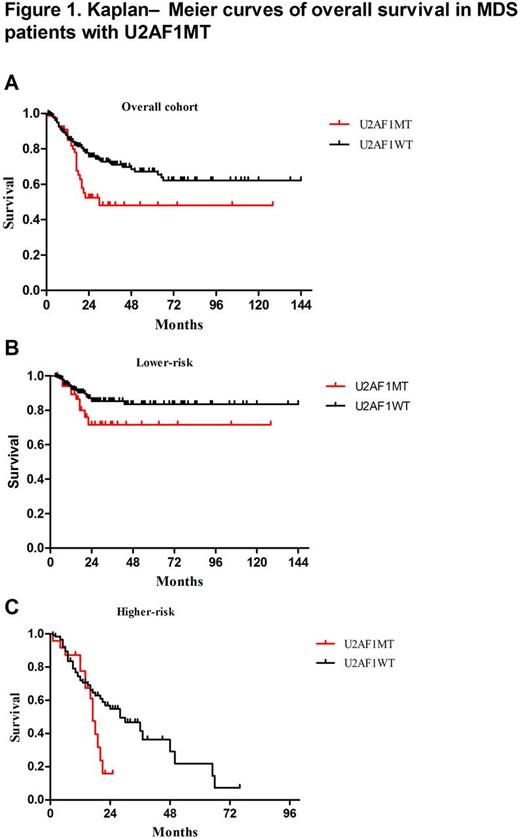
We identified U2AF1 MT in 86 (17%) subjects, including U2AF1S34F (64%, n=55), U2AF1S34Y (23%, n=20), U2AF1Q157R (6%, n=5), U2AF1Q157P (5%, n=4) and U2AF1R156H (2%, n=2). Compared to many MDS-related gene mutation, U2AF1 MT more frequently occurred in younger patients (Median age: 45 vs. 54 yrs; P=0.001). Compared to U2AF1 wildtype (U2AF1 WT ) subjects, U2AF1 MT subjects were significantly associated with anemia (Median HGB: 72 vs 78 g/L, P=0.015) and thrombocytopenia (Median PLT: 46 vs. 64×109/L, P=0.012). The U2AF1S34 subjects had more frequent platelet levels of <50×109/L (P=0.043) and the U2AF1Q157 or U2AF1R156 [HG1] subjects had more frequent hemoglobin concentrations at <80g/L ( P=0.008) with more overt fibrosis (P=0.049). We performed multivariate analysis to characterize the associations between U2AF1 MT and other common gene mutations and cytogenetic abnormalities, SF3B1 MT (P=0.001, OR=0.214, 95%CI: 0.083-0.55) was inversely associated with U2AF1 MT subjects, while isolated +8 (P<0.001, OR=4.671, 95%CI: 2.510-8.689), KRAS MT (P=0.008, OR=3.521, 95%CI: 1.388-8.934), and ASXL1 MT (P=0.033, OR=2.003, 95%CI: 1.059-3.786) was significantly enriched in U2AF1 MT subjects. Using copy number-adjusted VAF, we reconstructed the clonal architecture of U2AF1 MT patients to establish whether a U2AF1 MT was an ancestral or subclonal mutation. 65 patients with 1 or more mutation, except for U2AF1 MT, were analyzed. U2AF1 MT was an ancestral mutation in 46 (71%) patients and was a subclonal mutation in 19 (29%) patients. There was no significant association between ancestral- or subclonal U2AF1 MT and other gene mutations. In our cohort, U2AF1 MT was associated with worse overall survival (OS, P=0.01, Figure 1A). In subset analyses, we observed that U2AF1 MT tended to be associated with worse OS both in lower risk patients (P = 0.076, Figure 1B) and in higher risk patients (P = 0.064, Figure 1C). Patients with subclonal U2AF1 MT tended to have a reduced median OS than patients with ancestral U2AF1 MT (20 months vs. not reached, P = 0.369). Compared with ancestral U2AF1 MT patients, we found that subclonal U2AF1 MT tended to have a reduced median OS in lower-risk patients (22 vs. not reached, P= 0.211), but had an increased median OS in higher-risk patients (20 vs . 17 months, P = 0.54). Considering for different type of U2AF1 MT, MDS patients with U2AF1Q157/R156MT tended to have a reduced median survival as opposed to patients with S34 (17 vs . 30 months, P=0.318). Compared with U2AF1S34 MT patients, U2AF1Q157/R156 MT patients tended to have a worse prognosis in lower-risk patients (P=0.339), but not in higher-risk patients (P=0.989).
In summary, our study indicates that U2AF1 mutation is more common among younger MDS patients and represents as ancestral lesions in a substantial proportion of cases. MDS patients with a different type or different positions of U2AF1 mutations have distinct clinical and biological characteristics.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.