Key Points
Chronic HCV infection leads to extensive BCR immunoglobulin gene repertoire alterations with pathological features even in absence of MC.
Many large B-cell clones are consistently found, mainly among IgM+ memory B cells, showing a massive influence of HCV on this compartment.
Abstract
Patients chronically infected with hepatitis C virus (HCV) frequently develop mixed cryoglobulinemia (MC), a monoclonal expansion of immunoglobulin M (IgM)+ autoreactive B cells, and also have an increased B-cell lymphoma risk. Whether HCV infection also impacts the B-cell compartment and the B-cell receptor repertoire in patients not affected by MC or lymphomas is poorly understood. Flow cytometric analysis of peripheral blood B cells of 30 MC-negative HCV-infected patients and 15 healthy controls revealed that frequencies of class-switched memory B cells were increased in the patients, whereas frequencies of transitional and naive B cells were decreased. For 22 HCV+ patients and 7 healthy controls, we performed high-throughput sequencing of immunoglobulin heavy chain VDJ rearrangements of naive, mature CD5+, IgM+ memory, and class-switched memory B cells. An increased usage of several IGHV genes, including IGHV1-69 and IGHV4-59, which are closely linked to MC and HCV-associated lymphomas, was specifically seen among IgM+ memory B cells of the patients. Moreover, many, and partly very large, expanded clones were seen predominantly among IgM+ memory B cells of all HCV-infected patients analyzed. Thus, chronic HCV infection massively disturbs the B-cell compartment even in patients without clinically detectable B-cell lymphoproliferation and generates many large B-cell clones, especially among non–class-switched memory B cells. Because B-cell clones in MC and lymphomas derive from this B-cell subset, this establishes IgM+ memory B cells as a general target of lymphoproliferation in HCV+ patients, affecting apparently all patients.
Introduction
Chronic hepatitis C virus (HCV) infection not only causes manifestations in the liver but also frequently affects the B-cell compartment. The most common B-cell dysfunction, affecting up to 50% of HCV patients, is mixed cryoglobulinemia type II (MC), a disease characterized by monoclonal B-cell expansions producing immunoglobulin M (IgM) with rheumatoid factor activity.1 About 10% of HCV+ MC patients develop a B-cell non–Hodgkin lymphoma (NHL),2 but also patients without MC have an increased risk for an NHL.3-6 The most convincing evidence for a causative role of HCV in these lymphoproliferations is that its eradication upon antiviral therapy often results in resolution of NHLs and MC.7-9
The mechanism by which HCV triggers expansions of B-cell clones has not been fully elucidated. HCV may have a direct mutagenic effect on B cells10-15 and may also cause chronic antigenic stimulation of B cells expressing an HCV-specific B-cell receptor (BCR).16-19 A biased immunoglobulin heavy chain (IGH) repertoire with frequent usage of the IGHV1-69 and IGHV4-59 genes by B-cell clones of MC and HCV-associated NHLs supports this idea.16,20-24
Despite the occurrence of clonal expansions, the frequency of circulating B cells in HCV patients is not increased compared with healthy individuals,25,26 but B-cell subsets in HCV infection are apparently skewed, with increased transitional and activated memory B-cell frequencies.27,28 Accumulation of naive or CD5+ B cells has been reported in HCV-infected subjects.29,30 However, others reported reduced numbers of total and naive B cells in HCV patients with MC.31
Clonal B cells in HCV+ MC patients belong to the CD27+IgM+ (ie, non–class-switched [NCS]) memory B-cell subset and express somatically mutated immunoglobulins with rheumatoid factor activity.16,20 B-cell clones were also detected in some chronic HCV-infected patients in the absence of MC.32,33 However, it was not determined from which B-cell subsets these clones derived, and whether they preferentially used IGHV1-69. To date, the characterization of B-cell clonality in HCV infection has been based on analyses of bulk peripheral and/or intrahepatic B cells using conventional IGH VDJ gene polymerase chain reaction (PCR) approaches.34,35
Here, we analyzed the B-cell compartment in MC-negative HCV-infected patients for potential disturbances. In particular, we searched for (1) alterations in the B-cell subset composition, (2) potential biases in the BCR immunoglobulin gene repertoire of the main mature B-cell subsets, and (3) B-cell clonal expansions in specific B-cell subsets by high-throughput sequencing (HTSeq).
Materials and methods
Study participants
B cells were isolated from 40 mL peripheral blood collected from 30 treatment-naive patients with chronic HCV infection, attending the Department of Gastroenterology and Hepatology at the University Hospital Essen (supplemental Table 1, available on the Blood Web site). Patients were hepatitis B virus- and HIV-negative. Medical records were reviewed to exclude extrahepatic disease. Serum stored at 4°C in Wintrobe tubes for 7 days was used to assess the presence of cryoglobulins.36 For control, B cells were isolated from peripheral blood of 15 healthy donors. From an independent cohort of 29 treatment-naive HCV-positive patients, absolute numbers of CD19-positive B cells were determined by a flow cytometry–based quantification of lymphocyte subpopulations (Beckman Coulter, Krefeld, Germany) (supplemental Table 2). The study was approved by the local ethics committee of the University Hospital Essen, and written informed consent was obtained from all patients.
Cell sorting
B-cell subsets were isolated from peripheral blood after enrichment of CD19+ B cells with the MACS technology (Miltenyi Biotech, Bergisch Gladbach, Germany) on a FACSAria III cell sorter (Becton Dickinson, Heidelberg, Germany) upon 6-color staining for CD20, CD27, CD23, CD38, IgM, and CD5. Details are given in supplemental methods.
IGH VDJ rearrangement PCR
IGH repertoire analysis was performed on B cells from 22 HCV patients (supplemental Table 3) from the cohort of 30 patients, from whom at least 20 000 B cells were isolated per subset. DNA isolation was performed with the QiaAmp DNA micro kit (Qiagen, Hilden, Germany). To control for reproducibility of the results, and to aid identification of expanded clones, DNA from each B-cell population was split into 2 aliquots (replicates 1 and 2) of 50 ng DNA each (equivalent to 10 000 B cells), and IGH VDJ gene rearrangements of all IGHV subgroups were amplified, using the LymphoTrack IGH FR1 Assay kit (InvivoScribe, San Diego, CA). Amplified DNA was processed for HTSeq according to the LymphoTrack protocol. DNA was amplified with a 1-step multiplex PCR for 29 cycles using subgroup-specific IGH framework region 1 primers and IGHJ primers, incorporating platform-specific adaptors. PCR products were purified and pair-end sequenced using the MiSeq v3 reagent kit (Illumina, San Diego, CA). IGHV gene sequences have been deposited in the National Center for Biotechnology Information Short Reads Archive under accession number PRJNA418233.
Determination of clonality
Sequences with unidentified IGHV or IGHJ genes or CDR3 sequence were discarded. Sequences were considered clonally related when using the same IGHV and IGHJ genes, and having equal CDR3 nucleotide length with average nucleotide sequence identity of at least 90%, estimated with average linkage hierarchical clustering. To account for rare PCR-induced sequence variants, any 2 members of a clone need to have at least 2 nonshared substitutions, and at least a fivefold read coverage. Identical sequences were collapsed into 1 sequence. Only for the search of clonal expansions without substantial intraclonal diversity, we also considered the total number of reads for a given rearrangement. To be considered in the following steps of the analysis, each B-cell clone had to be discovered in both replicates. For further details, see supplemental methods.
Evaluation of HTSeq data and statistical analysis
The details of the computational processing and statistical evaluation of the HTSeq data are given in supplemental methods.
Results
Disturbance of B-cell subset composition in patients with chronic HCV infection
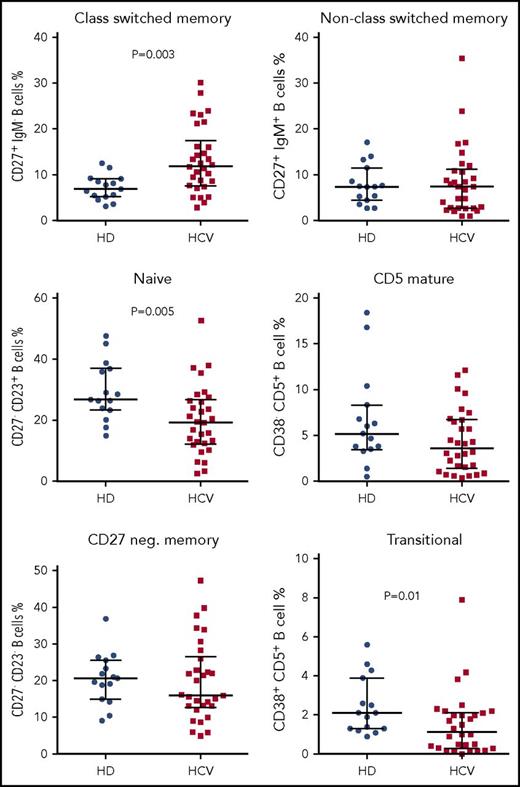
To investigate alterations of B-cell subset composition in patients with chronic HCV infection in the absence of MC or other lymphoproliferative diseases, we determined the percentages of the major peripheral B-cell subsets in 30 HCV-positive patients and 15 healthy blood donors by flow cytometry. CD20+ B cells were subdivided into naive, CD27+ class-switched (CS) memory, CD27+ NCS memory, mature CD5+, CD38+ transitional, and CD23−CD27− memory B cells (Figure 1). The frequency of CS memory B cells among CD20+ B cells was significantly increased in HCV patients as compared with healthy individuals (median, 12.0% vs 6.9%; P < .05). In contrast, naive and transitional B-cell frequencies were decreased in HCV patients compared with the control group (median, 19.4% vs 26.8% and 1.1% vs 2.1%, respectively; both comparisons with P < .05). No significant differences were observed for other subsets (Figure 2). As absolute numbers and frequencies of CD19+ B cells were not available for the first cohort, we estimated these values in an independent cohort of 29 HCV+ patients. Total number and frequency of peripheral blood CD19+ B cells of the HCV patients were in the range of values typical for healthy individuals (supplemental Table 2).
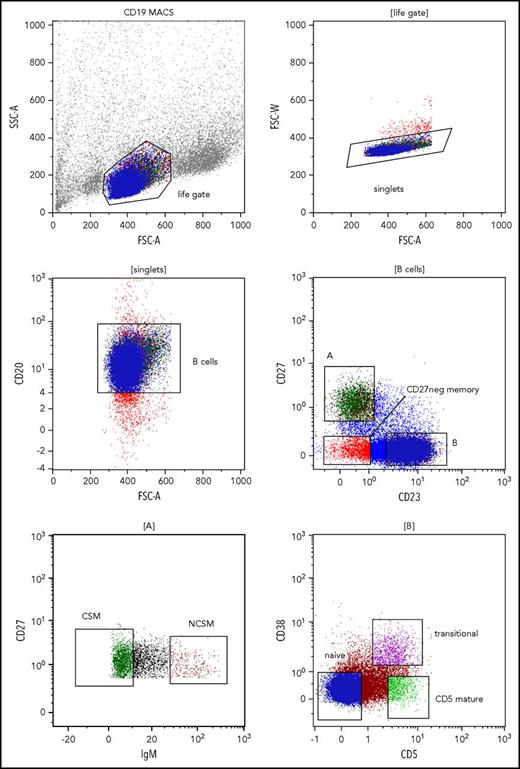
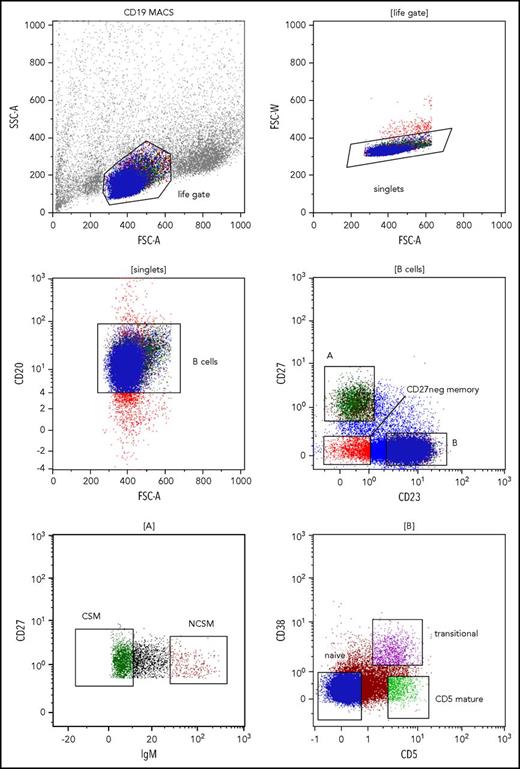
Gating strategies to identify peripheral blood B-cell subsets. Gating on lymphocytes was done according to forward scatter (FSC) vs side scatter area (SSC-A) features (upper left). Cell doublets were excluded by analyzing FSC area (FSC-A) vs FSC width (FSC-W) (upper right). In the next step, gating on CD20+ B cells was applied (middle left). CD27 vs CD23 staining was used to separate CD27+CD23− memory B cells (A) from CD23+CD27− naive, transitional, and mature CD5+ B cells (B), and from CD27−CD23− memory B cells (middle right). CD27+CD23− memory B cells were further subdivided into IgM-negative class-switched B cells (CSM) and IgM+ non–class-switched memory B cells (NCSM) (lower left). CD23+CD27− B cells were further separated into CD5−CD38− naive B cells, CD5+CD38+ transitional B cells, and CD5+CD38− mature CD5+ B cells (lower right). MACS, magnetic-activated cell sorting.
Gating strategies to identify peripheral blood B-cell subsets. Gating on lymphocytes was done according to forward scatter (FSC) vs side scatter area (SSC-A) features (upper left). Cell doublets were excluded by analyzing FSC area (FSC-A) vs FSC width (FSC-W) (upper right). In the next step, gating on CD20+ B cells was applied (middle left). CD27 vs CD23 staining was used to separate CD27+CD23− memory B cells (A) from CD23+CD27− naive, transitional, and mature CD5+ B cells (B), and from CD27−CD23− memory B cells (middle right). CD27+CD23− memory B cells were further subdivided into IgM-negative class-switched B cells (CSM) and IgM+ non–class-switched memory B cells (NCSM) (lower left). CD23+CD27− B cells were further separated into CD5−CD38− naive B cells, CD5+CD38+ transitional B cells, and CD5+CD38− mature CD5+ B cells (lower right). MACS, magnetic-activated cell sorting.
B-cell subset distribution in HCV patients compared with healthy individuals. B-cell subset frequencies were calculated on 30 HCV-positive patients and 15 healthy controls. HCV patients show an increase of CS memory B cells and a decrease of naive and transitional B cells, when compared with healthy individuals. Median with interquartile range and P values <.05 are shown. HD, healthy blood donors.
B-cell subset distribution in HCV patients compared with healthy individuals. B-cell subset frequencies were calculated on 30 HCV-positive patients and 15 healthy controls. HCV patients show an increase of CS memory B cells and a decrease of naive and transitional B cells, when compared with healthy individuals. Median with interquartile range and P values <.05 are shown. HD, healthy blood donors.
BCR immunoglobulin gene repertoire of peripheral blood B cells from HCV-infected individuals
To study potential BCR immunoglobulin gene repertoire disturbances and identify potential clonal expansions among mature B cells of HCV-infected patients not affected by MC, we isolated a minimum of 20 000 B cells per subset from the 4 main mature B-cell subsets, namely naive, mature CD5+, NCS memory, and CS memory B cells. Sufficient cell numbers were obtained from 22 patients and 7 healthy individuals. From each B-cell subset, 2 separate aliquots of DNA were prepared and used for amplification and HTSeq of IGH VDJ rearrangements. We obtained on average 484 207 processed reads per B-cell subset per individual (supplemental Table 4). Between 1600 and 19 200 unique productive (in frame) immunoglobulin VDJ rearrangements with at least 5 reads coverage were identified per B-cell subset per individual (average reads coverage: 68), corresponding to 78% to 90% of total VDJ sequences (not shown). For thorough quantification of the BCR immunoglobulin gene repertoire composition, only productive rearrangements were considered, because each productive rearrangement represents a single B cell.
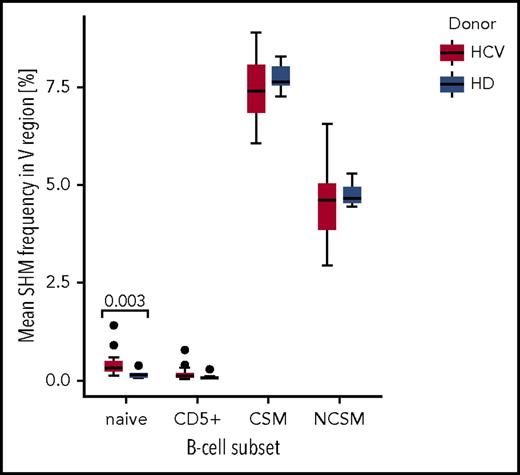
Somatic mutation analysis confirmed that naive and CD5+ B cells showed average mutation frequencies close to 0, in both healthy and HCV-infected individuals, with 2 outliers each in naive and CD5+ B cells of HCV patients (Figure 3). The memory B-cell subsets in HCV-infected patients harbored similar average mutation frequencies (7.4% for CS and 4.5% for NCS memory B cells) as healthy controls (7.8% for CS and 4.8% for NCS memory B cells) (Figure 3). Thus, somatic mutation levels among peripheral blood B-cell subsets do not differ between HCV patients and uninfected individuals, with the only differences seen for naive and CD5+ B cells in 2 HCV patients.
Comparison of mean somatic hypermutation (SHM) frequency in IGH VDJ sequences, estimated in the BCR immunoglobulin gene repertoires of 22 HCV-infected patients and 7 HD, across 4 B-cell subsets. Solid points represent outliers. As the 2 biological replicates for each B-cell subset were very similar, the mean of the 2 values was taken for the calculation. Only the IGHV genes of productive IGH VDJ rearrangements were considered for mutation analysis. The mean somatic mutation frequency estimated for each B-cell subset is for the healthy individuals: naive = 0.16%, CD5+ = 0.10%, CS memory = 7.76%, NCSM = 4.77%, and for the HCV+ patients: naive = 0.41%, CD5+ = 0.17%, CS memory = 7.43%, NCSM = 4.51%. For naive B cells, the P value (.003) indicates a significantly different mean SHM frequency between HCV patients and HD. The 2 outliers among naive and CD5+ B cells of HCV+ patients are for both B-cell subsets patients 11 and 12. In these cell samples, the higher average mutation load is due to a substantial number of rearrangements with mutation frequencies typical for memory B cells. Hence, the higher mutation load of these samples is likely due to contamination of the naive and CD5+ B cells by memory B cells, or in these patients, some NCSM B cells had reduced CD27 and increased CD23 expression, so that they were indistinguishable from the naive and CD5+ B cells in the sorting procedure. CD5+, CD5+ mature B cells.
Comparison of mean somatic hypermutation (SHM) frequency in IGH VDJ sequences, estimated in the BCR immunoglobulin gene repertoires of 22 HCV-infected patients and 7 HD, across 4 B-cell subsets. Solid points represent outliers. As the 2 biological replicates for each B-cell subset were very similar, the mean of the 2 values was taken for the calculation. Only the IGHV genes of productive IGH VDJ rearrangements were considered for mutation analysis. The mean somatic mutation frequency estimated for each B-cell subset is for the healthy individuals: naive = 0.16%, CD5+ = 0.10%, CS memory = 7.76%, NCSM = 4.77%, and for the HCV+ patients: naive = 0.41%, CD5+ = 0.17%, CS memory = 7.43%, NCSM = 4.51%. For naive B cells, the P value (.003) indicates a significantly different mean SHM frequency between HCV patients and HD. The 2 outliers among naive and CD5+ B cells of HCV+ patients are for both B-cell subsets patients 11 and 12. In these cell samples, the higher average mutation load is due to a substantial number of rearrangements with mutation frequencies typical for memory B cells. Hence, the higher mutation load of these samples is likely due to contamination of the naive and CD5+ B cells by memory B cells, or in these patients, some NCSM B cells had reduced CD27 and increased CD23 expression, so that they were indistinguishable from the naive and CD5+ B cells in the sorting procedure. CD5+, CD5+ mature B cells.
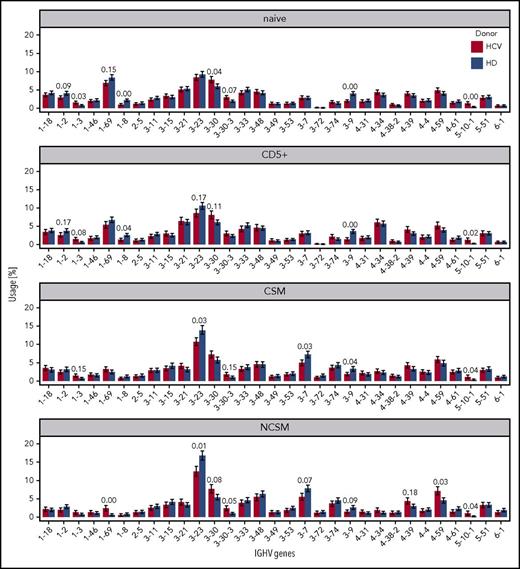
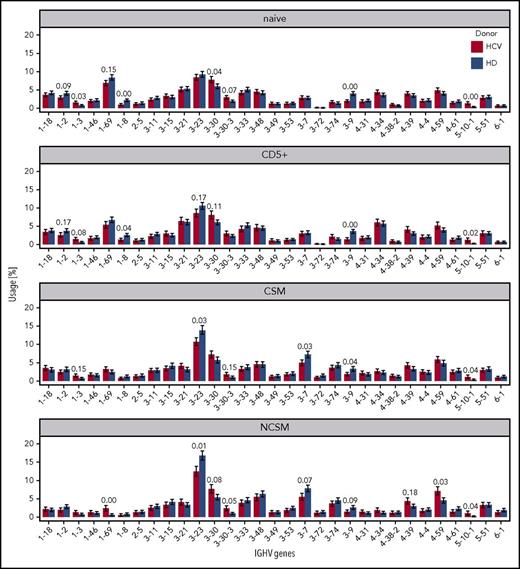
Next, we compared IGHV gene usage of B-cell subsets from HCV patients and healthy donors. All known potentially functional IGHV genes were found, confirming successful amplification of IGHV gene repertoires (not shown). For the analysis of IGHV gene usage, we focused on 31 IGHV genes used in at least 1 B-cell subset by at least 1% of cells (Figure 4; supplemental Figures 1 and 2). In each of the 4 B-cell subsets investigated, several IGHV genes showed a significantly higher or lower usage in HCV+ patients compared with healthy controls. For most IGHV genes with differential usage, no specific association with autoimmune diseases, lymphomas, and/or antigenic specificities is known. Strikingly, however, HCV-positive patients showed in the NCS memory B cells a higher usage of IGHV1-69, IGHV3-30, and IGHV4-59, that are known for their involvement in autoimmune diseases and/or in HCV-associated and other lymphomas.16,20-24 In conclusion, patients chronically infected by HCV show consistent disturbances in the usage of well-known disease-associated IGHV genes, mainly overrepresented among IgM memory B cells.
Relative use of individual IGHV genes in HCV+and healthy individuals, across the 4 B-cell subsets. Only those IGHV genes that accounted for at least 1% usage in at least 1 B-cell subset are shown. As a numerical measure to quantify the effect of HCV infection on IGHV segment usage, we computed for each IGHV segment the Bhattacharyya coefficient (BC) for posterior distributions of segment usage in HCV+ patients vs HD. BC values close to 0 (= 0 overlap between posteriors) indicate that there is a clear difference in the usage of the respective IGHV segment in HCV+ patients and HD. BC values close to 1 (= complete overlap of posteriors) mean that the usage of the respective segment is the same in HCV+ and healthy individuals. The 30 most significant differences between HCV+ patients and HD are annotated with BC. We also performed a further statistical evaluation of the data with the Mann-Whitney U test (supplemental Figure 2; supplemental methods). Whereas some IGHV genes showed a significant differential usage only with 1 or the other test, importantly, the IGHV genes IGHV1-69, IGHV3-30, and IGVH4-59 showed a significantly biased usage by NCSM B cells in HCV patients according to both tests.
Relative use of individual IGHV genes in HCV+and healthy individuals, across the 4 B-cell subsets. Only those IGHV genes that accounted for at least 1% usage in at least 1 B-cell subset are shown. As a numerical measure to quantify the effect of HCV infection on IGHV segment usage, we computed for each IGHV segment the Bhattacharyya coefficient (BC) for posterior distributions of segment usage in HCV+ patients vs HD. BC values close to 0 (= 0 overlap between posteriors) indicate that there is a clear difference in the usage of the respective IGHV segment in HCV+ patients and HD. BC values close to 1 (= complete overlap of posteriors) mean that the usage of the respective segment is the same in HCV+ and healthy individuals. The 30 most significant differences between HCV+ patients and HD are annotated with BC. We also performed a further statistical evaluation of the data with the Mann-Whitney U test (supplemental Figure 2; supplemental methods). Whereas some IGHV genes showed a significant differential usage only with 1 or the other test, importantly, the IGHV genes IGHV1-69, IGHV3-30, and IGVH4-59 showed a significantly biased usage by NCSM B cells in HCV patients according to both tests.
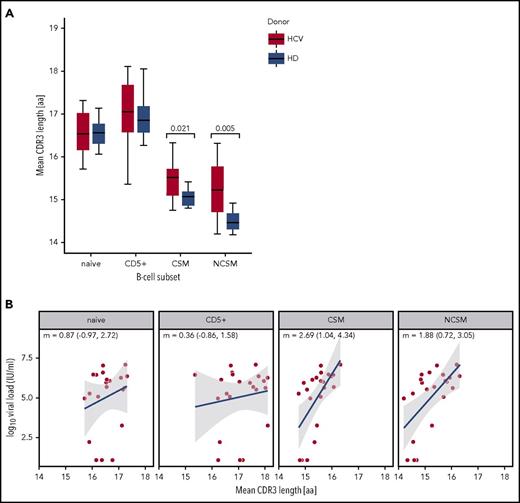
Alterations of the BCR immunoglobulin gene repertoire were also detectable for the average complementarity determining region 3 (CDR3) length of the productive VDJ gene rearrangements. Memory B cells have on average shorter CDR3s than naive B cells,37,38 as we also observed among cells from healthy and HCV-infected persons (Figure 5A). However, CS and NCS memory B cells from HCV+ patients had on average significantly longer mean CDR3 lengths than the respective populations from healthy individuals (Figure 5A). Notably, in memory B cells CDR3 lengths scale with the logarithm of viral load (Figure 5B). Hence, chronic HCV infection promotes the selection of unusually long heavy chain CDR3s in memory B cells.
Mean CDR3 length distribution and correlation with viral load. (A) Comparison of mean CDR3 sequence length in the BCR immunoglobulin gene repertoires of 22 HCV-infected patients and 7 healthy individuals (HD), across 4 B-cell subsets. As the 2 biological replicates for each B-cell subset were very similar, the mean of the 2 values was taken for the calculation. Only productive IGHV gene rearrangements were used for this analysis. The mean CDR3 sequence lengths estimated for each B-cell subset for the healthy donors are: naive = 16.51, CD5+ = 16.95, CSM = 15.05, NCSM = 14.51. For the HCV-infected patients, the values are: naive = 16.54, CD5+ = 17.07, CS memory = 15.47, NCSM = 15.24. Median with interquartile range and P values <.05 are shown (Mann-Whitney U test). (B) Linear relationship between log10-transformed viral load and mean CDR3 length in 22 HCV+ patients. Linear regression slopes (m) and 95% confidence intervals (CIs) for the slopes are shown for each B-cell subset. Blue best-fit lines with shaded 95% CIs for the regression lines are also shown. aa, amino acids.
Mean CDR3 length distribution and correlation with viral load. (A) Comparison of mean CDR3 sequence length in the BCR immunoglobulin gene repertoires of 22 HCV-infected patients and 7 healthy individuals (HD), across 4 B-cell subsets. As the 2 biological replicates for each B-cell subset were very similar, the mean of the 2 values was taken for the calculation. Only productive IGHV gene rearrangements were used for this analysis. The mean CDR3 sequence lengths estimated for each B-cell subset for the healthy donors are: naive = 16.51, CD5+ = 16.95, CSM = 15.05, NCSM = 14.51. For the HCV-infected patients, the values are: naive = 16.54, CD5+ = 17.07, CS memory = 15.47, NCSM = 15.24. Median with interquartile range and P values <.05 are shown (Mann-Whitney U test). (B) Linear relationship between log10-transformed viral load and mean CDR3 length in 22 HCV+ patients. Linear regression slopes (m) and 95% confidence intervals (CIs) for the slopes are shown for each B-cell subset. Blue best-fit lines with shaded 95% CIs for the regression lines are also shown. aa, amino acids.
Clonal expansions among B cells from MC-negative HCV-infected patients
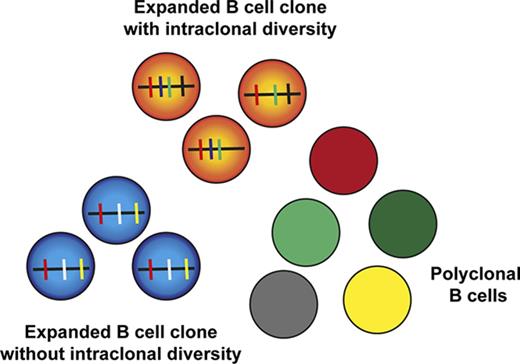
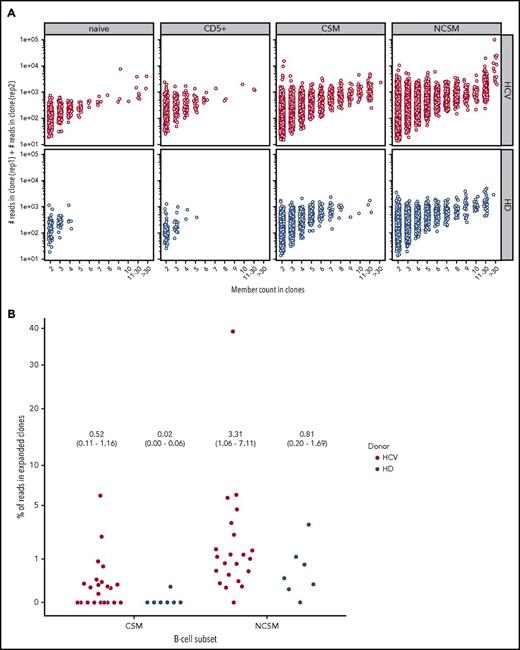
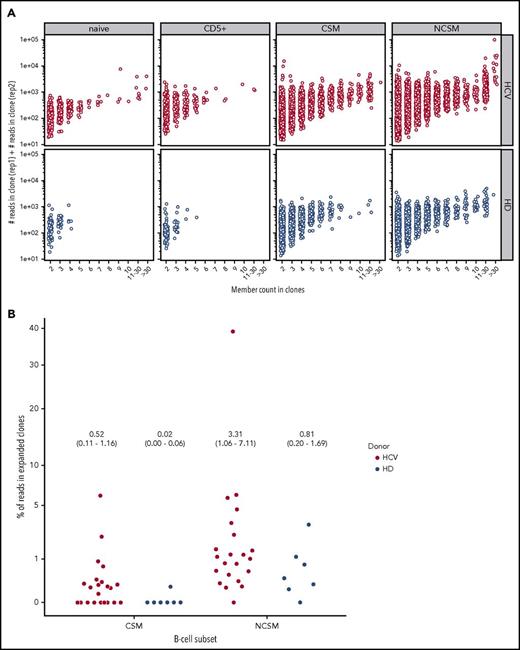
Given the large monoclonal B-cell expansions in HCV patients affected by MC, we wanted to assess whether HCV also affects the clonal composition of the main mature B-cell subsets in treatment-naive HCV-infected patients without MC. To reliably identify clonal expansions, we searched for sequences with the same IGHV and IGHJ genes, and highly similar CDR3 (equal CDR3 nucleotide length with at least 90% nucleotide sequence identity), and that differed from each other by at least 2 point mutations (or also 1, if both sequences were represented by a similar number of reads). Hence, if clone members with identical mutation patterns were present in a replicate, they were counted as 1. Moreover, substantially expanded B-cell clones should be found in both DNA replicates sequenced from each B-cell subset. These strict filtering criteria likely underestimate the number and sizes of clones. When using these criteria, hardly any clones were identified among naive and CD5+ B-cell subsets of healthy individuals; the few clones found had at most 3 or 4 members (Figure 6A). In contrast, among CS memory B cells from healthy donors, numerous clones with up to 8 members were detected, and among NCS memory B cells, there were even larger clones with up to 35 members (supplemental Table 5), in line with prior studies.37,39,40 Importantly, among NCS and CS memory B cells from chronically HCV-infected patients, a higher number of large clones with intraclonal diversity were identified (Figure 6A; supplemental Figure 3). Focusing on large clones with 10 or more members, overall, 20 of the 22 HCV+ patients harbored such clones among memory B cells. Seventeen patients harbored clonal expansions in NCS memory B cells with a total of 122 clones found, and 13 showed clonal expansions in CS memory B cells with a total of 35 clones (Table 1; supplemental Table 5). For comparison, of the 7 healthy individuals, 6 carried clonal expansions in NCS memory B cells with a total of 25 clones, and 1 individual in CS memory B cells with 1 clone. For both memory B-cell subsets, HCV patients showed on average more clones with at least 10 members than healthy individuals (Table 1). Moreover, considering large clones with >30 members, only 1 of the 25 clones among NCS memory B cells from healthy individuals had such a size, whereas in the NCS memory B cells from HCV-infected patients, 14 of the 122 B-cell clones consisted of >30 members (supplemental Table 5). There were few examples of large clones with members shared between B-cell subsets (see notes to supplemental Table 5).
Clonal-related B cells in HCV patients and healthy donors in the 4 B-cell subsets. (A) B-cell clones present in both replicates. Clones present in both replicates, shown as points according to their composition with x = clonal members and y = total read count. A higher concentration of larger clonal expansion can be observed in the NCSM B-cell subset of HCV-infected individuals. Indicated is the number of reads per clone member in both replicates. (B) The percent of reads in expanded clones relative to the total number of processed reads is shown for the 2 memory B-cell subsets for all individuals. The expanded clones considered here are the combination of clones with intraclonal diversity and at least 5 members in each replicate (supplemental Table 5) and the clones without substantial intraclonal diversity but an outlying number of reads (supplemental Table 6). The y-axis is shown on a “square-root scale.” In the figure, the average percent of reads in expanded clones per subset and per study group is given, with the 95% CI given in parentheses.
Clonal-related B cells in HCV patients and healthy donors in the 4 B-cell subsets. (A) B-cell clones present in both replicates. Clones present in both replicates, shown as points according to their composition with x = clonal members and y = total read count. A higher concentration of larger clonal expansion can be observed in the NCSM B-cell subset of HCV-infected individuals. Indicated is the number of reads per clone member in both replicates. (B) The percent of reads in expanded clones relative to the total number of processed reads is shown for the 2 memory B-cell subsets for all individuals. The expanded clones considered here are the combination of clones with intraclonal diversity and at least 5 members in each replicate (supplemental Table 5) and the clones without substantial intraclonal diversity but an outlying number of reads (supplemental Table 6). The y-axis is shown on a “square-root scale.” In the figure, the average percent of reads in expanded clones per subset and per study group is given, with the 95% CI given in parentheses.
Many clones in NCS memory B cells from HCV+ patients showed strong intraclonal diversification without mutations shared by all clone members, indicating that in these instances, naive B cells were driven into germinal center (GC) reactions and gave rise to diverse descendants from initiation of hypermutation onwards (supplemental Table 5; Figure 7). Other clones carried many shared mutations, meaning that an already mutated memory B cell likely underwent a secondary or tertiary GC reaction. Two exceptionally large clones, with >1000 and >300 members, were identified in the NCS memory subset from patients 23 and 31, respectively (supplemental Table 5). Strikingly, these 2 very large clones as well as 2 further among the 6 largest clones among NCS memory B cells expressed IGHV1-69. Overall, 8.2% and 7.4% of the 122 large clones among NCS memory B cells and 8.6% and 11.4% of the 35 CS memory B-cell clones from the HCV-infected patients used the IGHV1-69 or IGHV4-59 genes, respectively, whereas these genes were not found in the 26 large clones from healthy individuals. However, there was nevertheless a broad usage of IGHV genes among large expanded clones, with 27 distinct IGHV genes used by the clones among NCS memory B cells and 19 IGHV genes used by the clones among CS memory B cells (supplemental Table 5). The same 3 IGHV genes (IGHV3-7, IGHV3-23, IGHV3-48) were the most frequently used by expanded clones in NCS memory B cells of healthy individuals and HCV patients, indicating that for some IGHV genes the frequent usage by expanded clones in HCV patients is not a disease-associated feature but reflects a biased usage also seen in healthy adults.
Genealogic analysis of large memory B-cell clones with intraclonal diversity. (A-C) Selected lineages of expanded clones in the NCSM B-cell subset. (D-E) Selected lineages of expanded clones in the CS memory B-cell subset. (F) Selected lineage of the second largest clonal expansion discovered in this study in the NCSM B-cell subset. The grayscale indicates whether a given clonal member was found in replicate 1, in replicate 2, or in both (legend). The germline IGHV gene is given as the root of the tree, and any sequential mutation event between the germline and the clonal members is shown as a small black circle. The lineages belong to the following clones: (A) Pt1, NCSM, IGHV4-59_IGHJ4, members (replicate 1/replicate 2): 7/7; (B) Pt5, NCSM, IGHV3-43_IGHJ4, members: 8/7; (C) Pt12, NCSM, IGHV3-30_IGHJ4, members: 7/6; (D) Pt23, CSM, IGHV6-1_IGHJ6, members: 11/7; (E) Pt14, CSM, IGHV4-34_IGHJ5, members: 5/5; (F) Pt31, NCSM, IGHV1-69_IGHJ4, members: 185/137.
Genealogic analysis of large memory B-cell clones with intraclonal diversity. (A-C) Selected lineages of expanded clones in the NCSM B-cell subset. (D-E) Selected lineages of expanded clones in the CS memory B-cell subset. (F) Selected lineage of the second largest clonal expansion discovered in this study in the NCSM B-cell subset. The grayscale indicates whether a given clonal member was found in replicate 1, in replicate 2, or in both (legend). The germline IGHV gene is given as the root of the tree, and any sequential mutation event between the germline and the clonal members is shown as a small black circle. The lineages belong to the following clones: (A) Pt1, NCSM, IGHV4-59_IGHJ4, members (replicate 1/replicate 2): 7/7; (B) Pt5, NCSM, IGHV3-43_IGHJ4, members: 8/7; (C) Pt12, NCSM, IGHV3-30_IGHJ4, members: 7/6; (D) Pt23, CSM, IGHV6-1_IGHJ6, members: 11/7; (E) Pt14, CSM, IGHV4-34_IGHJ5, members: 5/5; (F) Pt31, NCSM, IGHV1-69_IGHJ4, members: 185/137.
A restriction of the approach used so far to identify B-cell clones is that it only detects clones with intraclonal diversification, because identical sequences in each aliquot were collapsed to a single sequence. To identify clonal expansions without substantial intraclonal diversification, we considered those clones that had only 1 to 2 distinct clonal members per replicate (identified because of >2 nucleotide differences) and were represented by an outlying (>700) number of reads, with the same VDJ rearrangements found in both replicates of a B-cell sample. Among NCS and CS memory B cells from HCV-infected patients, 41 and 8 rearrangements, respectively, were detected with 1500 to 15 000 total reads in both replicates (supplemental Table 6). These 49 clones, very likely representing expanded B-cell clones without substantial intraclonal diversity, were detected in 13 of the 22 HCV+ patients (supplemental Figure 3). Four of these clones used the IGHV4-59 gene, but none used IGHV1-69. Thirteen clones had unmutated VDJ sequences (<1% mutation frequency). In healthy individuals, only a single clone with so many sequencing reads was found among NCS memory B cells (supplemental Table 6). Consequently, for both memory B-cell subsets, the average number of such clones per individual was higher in HCV patients than healthy controls (Table 2).
Unexpectedly, both in healthy individuals and in HCV patients, few instances of clonally related B cells among naive B cells were found, with reads detected in both replicates of a sample. However, moderate homeostatic proliferation of human naive B cells has been described,41 and prior studies might have missed small clones among naive B cells, because identical or highly similar sequences are collapsed in HTSeq studies when only single samples per B-cell subset but no replicates are analyzed. Some of the identical sequences may also represent chance identity of rearrangements from independently generated cells or experimental artifacts, such as rare cell-sorting impurities (see note to supplemental Table 5).
Overall, we detected expanded B-cell clones in all HCV-infected patients (Figure 6A; supplemental Tables 5 and 6), mostly among NCS memory B cells. Indeed, when considering the fraction of reads assigned to expanded clones per individual in the memory B-cell subsets, the HCV patients showed higher average values (Figure 6B). Considering the consistent differences in terms of number and sizes of clones and their IGHV usage compared with healthy individuals, these clonal expansions clearly represent a HCV-induced phenomenon.
Discussion
Our knowledge of HCV and B-cell dysregulation has been mostly gained from studies of HCV-infected patients with MC. This study focused on chronic HCV carriers without extrahepatic manifestations. We found that CS memory B cells were significantly enriched and naive and transitional B cells reduced in HCV+ subjects compared with uninfected controls. There was, however, a wide variation in B-cell subset frequencies among patients (Figure 2). It remains unexplained why many patients show a higher frequency of CS but not of NCS memory B cells, because major alterations occur in the NCS memory cell compartment.
The analysis of somatic mutations of IGHV genes revealed no significant difference of mutation levels in the memory B-cell subsets from HCV+ subjects and uninfected controls, with values in expected ranges (Figure 3).38,39,42 Naive B cells and mature CD5+ B cells have, with rare exceptions, unmutated IGH VDJ rearrangements,43,44 and this was also observed in the HCV+ cohort. The few mutated cells detected might be due to rare cell-sorting impurities, to HCV-induced phenotypic changes of markers used for cell sorting, or to rare PCR errors in early PCR cycles (see legend to Figure 3). The higher mutation load of CS compared with NCS memory B cells is mainly due to the fact that NCS memory B cells are generated earlier in GC responses than CS memory B cells.39,40,44,45
IGHV1-69 and IGHV4-59 genes are frequently expressed in HCV-related B-cell disorders, namely MC B-cell clones producing rheumatoid factors and HCV-associated B-cell NHLs, and in other autoimmune diseases.18,20-24,46 Remarkably, these IGHV genes were more frequently used by NCS memory B cells of HCV-infected patients than those of healthy individuals (Figure 4). Thus, even in HCV patients without clinically detectable lymphoproliferation, there is a disease-associated disturbance of the BCR immunoglobulin gene repertoire, most pronounced in NCS memory B cells, with a significantly increased usage of those IGHV genes mainly involved in autoantibodies and pathological clonal B-cell expansions in HCV-infected patients.
Another major alteration of the IGH VDJ rearrangement structure in both the NCS and the CS memory B cells was detected for the CDR3 lengths of productive rearrangements. NCS and CS memory B cells from HCV-infected patients expressed on average longer CDR3s than the respective B-cell subsets of healthy humans (Figure 5A), with the CDR3 length scaling with the logarithm of viral load (Figure 5B). Given the biological function of CDR3 as antigen matching ligand, a rationale for this scaling could be that a linear increase in CDR3 length suffices to provide matching antibodies. Alternatively, but not mutually exclusive, the long CDR3s of memory B cells from HCV+ patients could also result from less efficient counterselection of cells with long CDR3s during memory B-cell generation as a consequence of HCV-mediated immune disturbances (with higher viral loads causing stronger immune alterations). This would also match the results of the IGHV gene usage analysis, which revealed a less efficient counterselection of genes frequently used by autoreactive B cells.
Within the memory B-cell compartment of healthy individuals, expanded memory B-cell clones can be detected.37,39,40 Therefore, to clearly distinguish any potential HCV-induced alterations of the B-cell clonal composition from physiological clonal expansions, it was critical to perform a parallel analysis of B cells from HCV patients and healthy controls. Because we studied 22 HCV patients, but only 7 healthy individuals, and considering the wide distribution of number and sizes of expanded clones among patients, the comparison in the 2 study groups for clones with and without substantial intraclonal diversity did not reach statistical significance (Tables 1 and 2; Figure 6B). However, large expanded B-cell clones were consistently more represented among the memory B cells of HCV patients than in the healthy controls. All patients harbored clones with and/or without intraclonal diversity. In particular, large clones (>10 members) with intraclonal diversity were detected in the NCS and CS memory B cells of 20 HCV patients with a total of 122 and 35 clones, respectively. Clonal expansions were also found in NCS and CS memory B cells of all the healthy controls, but at a lower frequency. Very large clones with >30 members were identified in NCS memory B cells of 5 HCV patients (24 clones overall), but only a single such clone was found in 1 healthy control (supplemental Table 5). A further disease-associated feature of the large clones among NCS memory B cells of HCV+ patients was the frequent usage of the IGHV1-69 or IGHV4-59 genes, which were not expressed in the large clones of healthy blood donors.
We also searched for clones without substantial intraclonal diversity. Again, clones were primarily detected in CS and NCS memory B cells of HCV-infected patients (Table 2). Whereas all clones from CS memory B cells of HCV+ patients were, as expected, somatically mutated, about a third of clones among NCS memory B cells of the patients were unmutated. The origin of these unmutated IgM+CD27+CD23− B cell clones is unclear, but may involve HCV-induced changes of B-cell marker expression on some B cells (see note to supplemental Table 6). However, some GC B-cell clones give rise to post-GC memory B cells even before somatic mutations are acquired.39,40 Some antigen-activated B cells receiving T-cell help may also become unmutated memory B cells without entering a GC.47
Taken together, we report a massive disturbance of the memory B-cell compartment in chronically HCV-infected patients even in the absence of clinically detectable lymphoproliferations. Overall, in all 22 patients investigated, we detected clonal expansions among memory B cells that, regarding frequency and size, are clearly distinguished from physiological memory B-cell clones. Many of these clones have intraclonal diversity, a feature of diversification in GC reactions, where proliferating clones undergo somatic hypermutation. However, we also detected numerous clones without substantial intraclonal diversification most likely derived from a post-GC proliferation. The detection of increased usage of MC-, autoantibody-, and HCV lymphoma–associated IGHV genes by the NCS memory B cells in general, and especially in the large clones, revealed that this is a general phenomenon in HCV+ patients. Finally, the strong disturbances seen among NCS memory B cells highly resemble lymphoproliferative diseases in HCV patients, because MC clones and HCV-associated B-cell lymphomas are mostly IgM+ and somatically mutated (ie, are linked to NCS memory B cells).16-20 The reasons for the preferential involvement of NCS memory B cells in the clonal expansions are not well understood. However, IgM+ memory B cells apparently have a higher propensity to undergo proliferation (and repeated GC reactions) upon restimulation than CS memory B cells, which prefer a plasma cell differentiation program.45,48 There might thus be a selective pressure on these cells to remain unswitched. It remains to be clarified by which mechanisms expanded IgM+ B-cell clones in initially asymptomatic HCV-infected patients become MC clones. Several factors might contribute to this, such as expression of HLA allotypes linked to an increased risk of MC development, and/or an antigen-dependent affinity maturation process that increases the rheumatoid factor activity of IgMs, or the acquisition of some genetic lesions in expanded B-cell clones.49,50
The data reported in this article have been deposited in the National Center for Biotechnology Information Short Reads Archive database (accession number PRJNA418233).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kerstin Heise for expert technical assistance, and Marc Seifert for critical reading of the manuscript. They also thank Klaus Lennartz and the Imaging Center Essen (IMCES) core facility for support in cell sorting, and Bettina Budeus for support in the bioinformatic analysis.
This work was supported by the Deutsche Forschungsgemeinschaft through TRR60 (R.K., D.H., and J.D.), and by the Dr Werner Jackstädt-Stiftung (P.J. and R.K.).
Authorship
Contribution: F.A.T., J.D., and R.K. designed the study; F.A.T. performed the research; S.K. and D.H. performed the bioinformatic evaluation; A.K. provided essential blood samples and clinical data; L.K.-H. performed the HTSeq analysis; P.J. collected clinical data and contributed to project coordination; and F.A.T. and R.K. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ralf Küppers, Institute of Cell Biology (Cancer Research), University of Duisburg-Essen, Medical Faculty, Virchowstr 173, 45122 Essen, Germany; e-mail: ralf.kueppers@uk-essen.de.