In this issue of Blood, Ivaldi et al showed that the BGLT3 long noncoding RNA (lncRNA) locus increases the production of γ-globin, the fetal form of β-globin.1 The BGLT3 transcript itself plays a role, and the locus displays enhancer-like features.
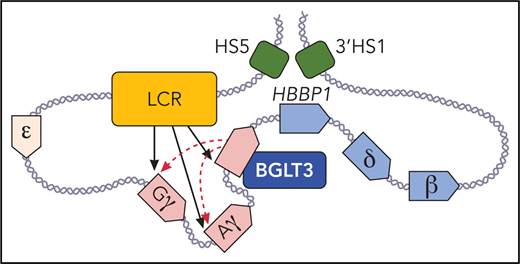
The long noncoding gene BGLT3 helps drive fetal γ-globin expression via its RNA transcript and an enhancer-like activity. BGLT3 contacts the γ-globin promoters and the locus control region (LCR; shown in orange). Fetal expressed γ-globin and BGLT3 genes are depicted in red, and adult expressed δ- and β-globin genes and HBBP1 are shown in blue. DNAse I hypersensitive sites (HS) 5, and 3′ HS1 (in green) that flank the β-globin locus are in physical proximity with the HBBP1 noncoding gene in fetal erythroblasts. Professional illustration by Patrick Lane, ScEYEnce Studios.
The long noncoding gene BGLT3 helps drive fetal γ-globin expression via its RNA transcript and an enhancer-like activity. BGLT3 contacts the γ-globin promoters and the locus control region (LCR; shown in orange). Fetal expressed γ-globin and BGLT3 genes are depicted in red, and adult expressed δ- and β-globin genes and HBBP1 are shown in blue. DNAse I hypersensitive sites (HS) 5, and 3′ HS1 (in green) that flank the β-globin locus are in physical proximity with the HBBP1 noncoding gene in fetal erythroblasts. Professional illustration by Patrick Lane, ScEYEnce Studios.
This observation is of considerable interest because boosting the expression of fetal globin alleviates the symptoms of the inherited blood disorders sickle cell disease and β-thalassemia.2 Those aiming to increase fetal globin expression by using CRISPR or TALENs to edit out repressor binding sites in blood stem cells may wish to avoid perturbing this new activating element, and researchers who are investigating fundamental mechanisms of gene expression will note that BLGT3 is an example of a lncRNA gene that contributes to gene activation.
The BGLT3 gene lies downstream of the duplicated γ-globin genes and upstream of the adult δ-globin and β-globin genes in a region termed the intergenic region (see figure). This region was once like the Bermuda Triangle of the β-globin locus, with researchers drifting into it and sometimes failing to emerge with clear answers.
Early work showed that naturally occurring deletions in the region led to hereditary persistence of fetal hemoglobin, a benign condition in which the fetal globin genes continue to be expressed throughout life, rather than being silenced at birth when the adult globin genes take over.3 It was thought that the deletions either removed repressive elements or juxtaposed downstream enhancers close to the fetal globin genes. But results from transgenic mice or gene editing in human cells did not always recapitulate the human phenotypes.4,5 It now seems that the intergenic region has additional important features, most notably the 2 genes that produce noncoding RNA, BGLT3 (discussed by Ivaldi et al) and the pseudogene HBBP1.6
Ivaldi et al map the BGLT3 transcript and show that it is coexpressed with the fetal globin genes during erythroid development and maturation. BGLT3 also comes into physical proximity with the γ-globin genes via chromatin looping. Deleting the BGLT3 gene or depleting its transcript via a dCas9-imposed elongation block or by antisense oligo nucleotides in K562 cells diminished γ-globin expression, suggesting an activating role of BGLT3. These are notable findings because studies on numerous lncRNAs, including in those erythroid cells, have sometimes failed to reveal clear functions.7
Interestingly, the act of transcription elongation but not the transcript per se seem to be required for the looped contacts, assuming that the BGLT3 transcripts were sufficiently depleted in the nuclei. Forced expression of BGLT3 modestly upregulated fetal globin expression in some contexts, but the RNA is not potent when provided in trans. Ivaldi et al observed that the region encompassing the BGLT3 gene has the hallmarks of an enhancer, which is seemingly held in an active state via transcription.
The function of BGLT3 contrasts with the other noncoding gene, HBBP1, which resides downstream of BGLT3 and is expressed only in adult erythroid cells.6 There, Capture-C showed that HBBP1 and BGLT3 demarcate not only a functional separation of fetal vs adult gene expression, but they also reside in sharply separated developmental stage-specific chromatin contact domains. HBBP1 contributes to fetal globin repression, possibly by binding the repressor BCL11A, which suggests that it might be a good candidate for CRISPR-mediated deletion aimed at boosting fetal globin expression. In contrast to BGLT3, the HBBP1 gene region, and not the transcript itself or the act of transcription through the gene, is required for influencing the architecture of the locus, presumably by separating the fetal or adult globin genes from the powerful locus control region (LCR) enhancer at different developmental stages.6 Thus, 2 adjacent noncoding genes function by different mechanisms to facilitate globin switching.
Ivaldi et al propose that BGLT3 operates via multiple mechanisms, including maintaining an enhancer-like state, looping to the γ-globin genes, and recruiting co-regulators such as the mediator complex possibly via the BGLT3 transcript itself. Other transcripts in this region, such as the noncoding RNA HIDALGO,8 have previously been shown to activate the fetal globin gene, so a solid foundation has been laid, and future work will likely map these mechanisms or chart their interrelatedness. Aficionados will also be interested in investigating why BGLT3 loops not only to the γ-globin genes but also to the LCR and why only the γ-globin interaction is sensitive to inhibition of BGLT3 transcription. With increasingly efficient genome and epigenome editing technologies, this very carefully executed study could soon be complemented by experimental perturbations in primary human fetal erythroid cells.
The mysteries of this region are gradually being resolved, but the full regulation of the locus remains incompletely understood. The reductionist approach of studying one element at a time has been effective here and in the past. It is also notable that other noncoding RNAs emerging from the LCR itself,9 and multiple additional cis elements implicated by natural point mutations, deletions, or in mapping experiments also have an effect and have presumably evolved together to confer a complex and robust pattern of gene expression on the locus.2
Not surprisingly, given that regulatory elements can work across long distances and that multiple elements can feed into a system to determine the ultimate expression pattern and level, multiple components influence globin gene expression. Understanding all of them is challenging, but disrupting individual elements to alleviate fetal globin repression may be possible without understanding everything. The more we know, the more likely we are to be able to choose and introduce targeted alterations that boost fetal globin efficiently while not interfering with the expression of other genes in the genome.
Conflict-of-interest disclosure: The authors declare no competing financial interests.