Abstract
Introduction: Acute Myeloid Leukemia (AML) is a heterogenous malignancy with the most common genomic alterations including mutations in NPM1, DNMT3A, and FLT3. Multiple FLT3-inhibitors have been developed and Midostaurin is the first to be FDA approved. However, while Midostaurin is now part of standard of care for FLT3 mutant AML patients undergoing induction chemotherapy, there is still a spectrum of clinical response. We hypothesized that additional biological factors could play a role in this variation of response. We previously observed that FLT3-ITD wildtype samples with KRAS mutations correlated with increased resistance to Midostaurin (1). Here, we utilized the Beat AML dataset containing over 900 AML patient samples processed through an ex vivo drug sensitivity screen to understand further whether genomic alterations could influence the range of Midostaurin response.
Methods:We identified 180 (46 FLT3-ITD positive, 134 FLT3-ITD negative) de-novo primary AML peripheral blood or bone marrow samples from distinct patients within the Beat AML dataset. All samples included RNA-Sequencing profiling, mutational analyses, and ex-vivo Midostaurin drug screening data. Drug sensitivity was measured as area under a seven-point drug concentration curve (AUC). AUC was calculated as the area under the fitted probit curve (via direct integration) using all seven dose ranges as x-values and cell viability with limits from 0 to 100% as the y-value. RNA-Sequencing was normalized to counts per million (cpm) and the overall gene expression was chosen as the transcript of that gene with greatest average expression across the cohort. Differential gene expression was performed with the EdgeR package in R version 3.4.0.
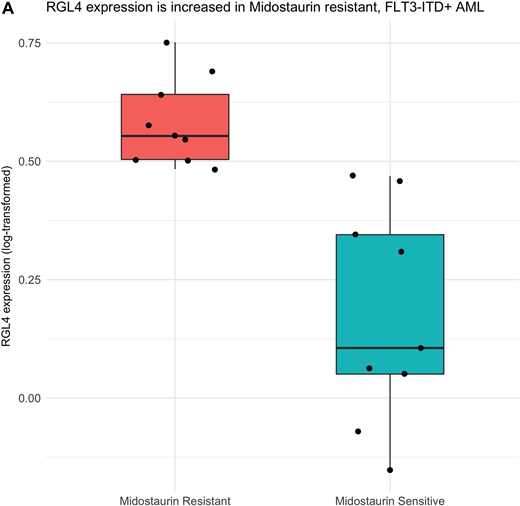
Results: As expected FLT3-ITD mutant samples, on average, are more sensitive to Midostaurin than FLT3-ITD wildtype sample. However, within the FLT3-ITD mutant cohort we observed a distribution of sensitivity responses (AUC mean of 55.4, IQR 35.5 - 75.3). Analysis of differential gene expression performed on the top 20% and bottom 20% of AUC values identified RGL4 over-expression across the entire resistant cohort (n = 9; Figure A). This was validated in an orthogonal data-set of 79 samples (21 mutant FLT3-ITD, 58 wildtype FLT3-ITD). To gain even more power within the validation set, we looked at all FLT3-ITD positive AUC values and observed a positive correlation between Midostaurin AUC and RGL4 expression (Spearman's rank correlation coefficient of 0.72, p < 0.05).
Conclusions: In summary, we found that RGL4 over-expression correlated with resistance to Midostaurin in FLT3-ITD mutant samples in our ex vivo drug screen. RGL4 (ral guanine nucleotide dissociation stimulator like 4) encodes a protein similar to the guanine nucleotide exchange factor for Ral known as Ral GDS (guanine dissociation stimulator). It is highly expressed in the bone marrow and has the potential to activate the Ras-Raf-MEK-ERK pathway. As we had previously observed KRAS mutations correlating with resistance in FLT3-WT samples, this additional finding of RGL4 over expression supports the involvement of RAS pathway activation as a potential mechanism of resistance to Midostaurin. Additional in vitro studies are necessary to establish and further understand this mechanism as well as to test the efficacy of a Midostaurin / MEK inhibitor combination to treat resistant samples.
1. Watanabe-Smith, K., Rosenberg, M., Bucy, T., Tyner, J. W., & Borate, U.(2017). Factors Predicting Response and Resistance to Midostaurin in FLT3 Positive and FLT3 Negative AML in 483 Primary AML Patient Samples. Blood,130(Suppl 1), 296.
Borate:Novartis: Consultancy; Agios: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.