In this issue of Blood, Kurotaki et al demonstrate that IRF8 expression delineates a subset of lymphoid-primed multipotent progenitors (LMPPs) biased toward the generation of conventional type 1 dendritic cells (cDC1s) by epigenetically priming the precursors for the expression of dendritic cell (DC) lineage genes.1
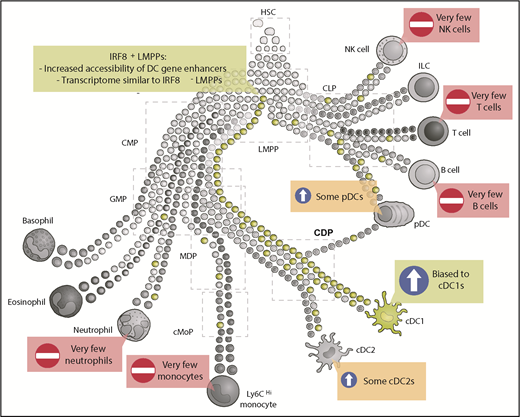
IRF8+ LMPPs are biased toward cDC1 differentiation. Schematic representation of hematopoietic development where some early progenitors are already precommitted to certain lineages (adapted from Guilliams et al5 ). Expression of IRF8 in LMPPs (green) biases differentiation toward cDC1s. Some IRF8+ LMPPs also give rise to cDC2s and pDCs, whereas a few IRF8+ LMPPs will give rise to other lineages, including monocytes, neutrophils, T cells, or B cells. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; HSC, hematopoietic stem cell; ILC, innate lymphoid cell; NK, natural killer.
IRF8+ LMPPs are biased toward cDC1 differentiation. Schematic representation of hematopoietic development where some early progenitors are already precommitted to certain lineages (adapted from Guilliams et al5 ). Expression of IRF8 in LMPPs (green) biases differentiation toward cDC1s. Some IRF8+ LMPPs also give rise to cDC2s and pDCs, whereas a few IRF8+ LMPPs will give rise to other lineages, including monocytes, neutrophils, T cells, or B cells. CLP, common lymphoid progenitor; CMP, common myeloid progenitor; HSC, hematopoietic stem cell; ILC, innate lymphoid cell; NK, natural killer.
cDC1s are a conventional DC subset specialized in the cross-presentation of intracellular antigens. cDC1s develop in a stepwise manner from hematopoietic stem cells to LMPPs, common myeloid progenitors, granulocyte macrophage progenitors (GMPs), common dendritic cell progenitors (CDPs), pre-cDC1s, and, finally, to cDC1s. At each stage of development, multipotent progenitors (MPPs) are thought to make a decision to progress to a more restricted progenitor and, eventually, a specific cell lineage. However, the idea of a true MPP has recently been questioned, with cellular barcoding and single-cell analyses suggesting that some LMPPs may be preprogrammed toward a specific lineage.2-5
Interferon regulatory factor 8 (IRF8) has long been associated with cDC1 development and maintenance. We recently proposed IRF8 to be a “terminal selector” for cDC1s, a transcription factor (TF) whose expression is continuously required to prevent loss of identity and death.6 IRF8 is gradually upregulated during the different stages of cDC1 development, and its expression has been shown to be induced and maintained via autoactivation by the TFs PU.1 and Batf3.7
In this study, Kurotaki et al build on their previous work understanding the role of IRF8 in mononuclear phagocyte biology, identifying a subset of IRF8-expressing LMPPs that are biased toward the cDC1 lineage. Mechanistically, they show that this bias toward cDC1s is not the result of an altered transcriptome in IRF8+ LMPPs compared with their IRF8− counterparts. Rather, the acquisition of IRF8 expression results in epigenetic changes in the LMPPs, leading to chromatin around genes associated with the DC lineage being made accessible and, thus, later leading to the transcriptional profile observed in DC committed progenitors and DCs themselves but not the earlier upstream progenitors, GMPs, or common monocyte progenitors (cMoPs).1 Importantly, IRF8 itself seems crucial for introducing this bias because PU.1-IRF8 composite binding motifs were found to be enriched in the assay for transposase-accessible chromatin with high-throughput sequencing (ATAC-Seq) peaks present in the IRF8-expressing LMPPs but not in the IRF8− LMPPs. Moreover, these epigenetic changes were absent in IRF8-null LMPPs; although formal proof for direct IRF8 involvement will come from IRF8 chromatin immunoprecipitation on LMPPs, this was confirmed in previously published data on monocyte/DC progenitors (MDPs).1 Underlining the translational value of these findings, this report is in line with a recent study in humans in which expression of IRF8 in early MPPs marked cells biased toward the DC lineage.4
Several key questions arise based on these findings. First, do all cDC1s arise through an IRF8+ LMPP? Kurotaki et al demonstrate that IRF8− LMPPs also retain the capacity to generate cDC1s, although this appears slower than observed with IRF8+ LMPPs. They also show that at least some IRF8− LMPPs can acquire IRF8 expression. Perhaps this is the cause of the delayed development, in that they may need to acquire IRF8 expression first. However, alternatively, one could also envisage distinct pathways for cDC1 development: some IRF8+ precursors may preferentially become cDC1s, and some precursors may acquire IRF8 later and represent true MPPs, or perhaps precursors preferentially programmed for another lineage can still switch toward cDC1s relatively late. A second question pertaining to this is then, why does IRF8 expression in the LMPP specify cells toward the cDC1 lineage when MDPs, CDPs, and cMoPs also express IRF8 and these are not all biased toward cDC1s? In fact, IRF8 also controls a set of enhancers highly expressed in monocytes,8 and loss of IRF8 impedes the progression from cMoPs to monocytes.6 Thus, how does IRF8 safeguard differentiation toward other immune lineages at these later time points? Perhaps through cooperation with other TFs? Indeed, Kurotaki et al demonstrated that expression of the TF hepatic leukemia factor can skew LMPPs away from the cDC1 lineage and more toward cDC2s. The interaction with PU.1 is also important to consider, because PU.1-IRF8 composite binding motifs are (as for IRF8+ LMPPs) enriched during the progression from the GMP to MDP8 and, hence, could be essential for safeguarding cDC1 and monocyte fate. Further work will be required to understand how IRF8 interacts with other TFs at the different stages of development to induce specification toward distinct myeloid cell lineages.
Additionally, although there was a significant bias within the IRF8-expressing LMPPs toward generating cDC1s, this was not the only differentiation outcome: some plasmacytoid DCs (pDCs) and cDC2s were also generated, as well as low numbers of neutrophils and monocytes1 (see figure). Because the ATAC-Seq experiments identifying the altered chromatin states were performed as bulk analyses, it remains an open question whether the epigenetic landscape is the same within those IRF8-expressing LMPPs that give rise to cDC1s compared with the IRF8-expressing LMPPs that will go on to generate cDC2s, pDCs, or monocytes.
Finally, how is IRF8 induced in the LMPPs? Interferon-γ (IFN-γ) signaling represents a potential candidate.9 Interestingly, because the investigators speculate that IRF8-expressing LMPPs could represent a “priority lane” to generating cDC1s, IFN-induced cDC1 development could represent a positive-feedback loop developed in response to evolutionary pressure from intracellular pathogens. It has been proposed that, following infection with toxoplasma, interleukin-12 production by peripheral cDC1s induces the production of IFN-γ by natural killer cells in the bone marrow.10 Thus, this IFN-γ might drive the expression of IRF8 in LMPPs, promoting the accelerated biased development toward cDC1s.
Thus, this report highlights a population of LMPPs expressing IRF8 that is biased toward cDC1 differentiation as the result of an altered epigenetic landscape. This highlights the importance of epigenetics and not just transcriptomics when studying cell fates and further expands recent work suggesting that we need to reconsider the nature of true MPPs.
Conflict-of-interest disclosure: The authors declare no competing financial interests.