Both founder and secondary mutations in AML and MDS have prognostic implications that may depend on their rank in subclonal hierarchy. Mutational profiles at relapse after chemotherapy may be different from relapse following alloHCT as a result of the graft-versus-leukemia (GVL) effect. We hypothesized that the molecular landscape of AML and MDS at the time of post-alloHCT relapse may (1) recapitulate the clonal hierarchy of the founder and secondary mutations at diagnosis, (2) be represented by founder mutations without secondary mutations, or (3) be characterized by acquisition of new mutations.
Using bone marrow samples banked at diagnosis and at the time of relapse, we compared mutational characteristics using a targeted multi-amplicon deep NGS panel of the 176 most commonly mutated genes in myeloid neoplasia. Thirty patients (pts) with AML (n=22) or MDS (n=8) who relapsed after their first alloHCT were included as the "transplant group", and samples from diagnosis or pre-HCT relapse (if available), whichever was last, and at relapse were compared. A comparison "chemotherapy group" included 11 pts with AML (n=7) or MDS (n=4) who relapsed after achieving complete remission (CR) from chemotherapy were included to compare mutational changes from diagnosis to relapse. For this analysis, we selected the 27 most commonly observed somatic mutations.
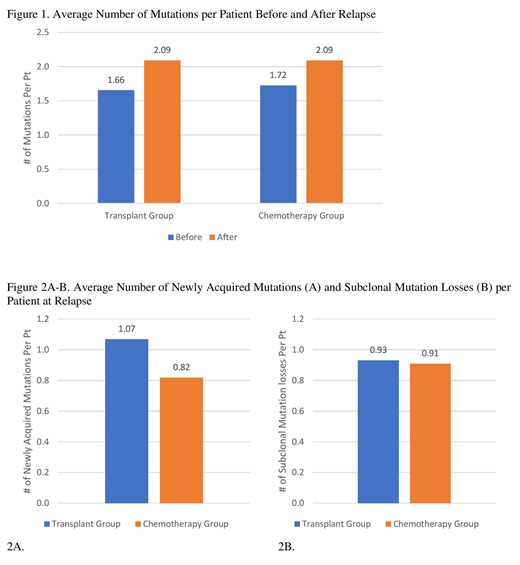
For the transplant group, median age at alloHCT was 59 years (range 32-73). Cytogenetics at diagnosis were normal in 47% and complex in 20%. At diagnosis, IDH2, NPM1, ASXL1, RUNX1, FLT3, and TP53 mutations were found in 13%, 13%, 10%, 10%, 7%, and 3% of pts, respectively. The majority received reduced-intensity conditioning (53%), bone marrow grafts (53%) from matched unrelated donors (55%). Sixteen patients (AML=15, MDS=1) were in CR at transplant. Median time from alloHCT to relapse was 114 days (median 28-935 days). The mutational profiles for all patients changed at post-alloHCT relapse. While the majority both gained and lost different mutations (60%), some just gained (27%) or lost mutations (13%) at relapse. The number of mutations gained ranged from 0-5 (median 1) and mutations disappeared ranged from 0-5 (median 2). The most common newly acquired mutation was FLT3 (17%) followed by DNMT3A (13%). Two cases (7%) showed re-acquisition of mutations that were present at diagnosis but disappearing after induction; 1 patient re-acquired FLT3, NPM1, and RIT1 mutations while another reacquired a TET2 mutation. Most commonly disappearing mutations at post-alloHCT relapse was IDH2 (10%). DNMT3A (20%) was the most common mutation that persisted through chemotherapy and alloHCT.
For the chemotherapy group, median age at diagnosis was 59 (range 32-71). Cytogenetics at diagnosis were normal in 55% and complex in 18%. At diagnosis, NPM1, FLT3, TP53, and ASXL1, mutations were found in 27%, 18%, 18%, and 9%, respectively; none had IDH2 or RUNX1 mutations. Median time from diagnosis to relapse was 154 days (range 33-1,716 days). As in the post-alloHCT group, changes in the mutational profile were found in all patients. Six patients (55%) gained newly acquired mutations while 5 patients lost a mutation without acquiring a new mutation: 2 cases with TP53 loss and 1 case each of PTPN11, RIT1, and TET2 losses. The number of mutations gained and disappeared per patient varied (each median 1, range 0-3). The most commonly gained mutations were DNMT3A and TET2 (18% each). DNMT3A and GNAS were the most commonly persistent mutations at chemotherapy relapse (18% each).
The biology of relapse following alloHCT is complex. Our results show that while clonal changes were always present at relapse following chemotherapy or following alloHCT, the mutational evolution was more diverse and complex at post-alloHCT relapse as compared to relapse after chemotherapy alone. This results signifies the importance of mutational profile test at relapse for the targeted therapy. Larger scale analyses can further elucidate somatic hits suggesting the mechanism of escape from the GVL.
Gerds:Incyte: Consultancy, Research Funding; Pfizer: Consultancy; Celgene Corporation: Consultancy, Research Funding; CTI Biopharma: Consultancy, Research Funding; Roche: Research Funding; Sierra Oncology: Research Funding; Imago Biosciences: Research Funding. Sekeres:Syros: Membership on an entity's Board of Directors or advisory committees; Celgene: Membership on an entity's Board of Directors or advisory committees; Millenium: Membership on an entity's Board of Directors or advisory committees. Mukherjee:McGraw Hill Hematology Oncology Board Review: Other: Editor; Pfizer: Honoraria; Novartis: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Bristol-Myers Squibb: Speakers Bureau; Takeda: Membership on an entity's Board of Directors or advisory committees; Projects in Knowledge: Honoraria; Celgene Corporation: Consultancy, Membership on an entity's Board of Directors or advisory committees, Research Funding; Partnership for Health Analytic Research, LLC (PHAR, LLC): Consultancy. Advani:Glycomimetics: Consultancy, Research Funding; Abbvie: Research Funding; Macrogenics: Research Funding; Pfizer: Honoraria, Research Funding; Kite Pharmaceuticals: Consultancy; Amgen: Research Funding. Nazha:Jazz Pharmacutical: Research Funding; Abbvie: Consultancy; Daiichi Sankyo: Consultancy; MEI: Other: Data monitoring Committee; Tolero, Karyopharma: Honoraria; Incyte: Speakers Bureau; Novartis: Speakers Bureau. Majhail:Mallinckrodt: Honoraria; Nkarta: Consultancy; Anthem, Inc.: Consultancy; Incyte: Consultancy; Atara Bio: Consultancy. Maciejewski:Novartis: Consultancy; Alexion: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.