Introduction: New approaches to find and then drug pediatric acute myeloid leukemia (AML)-specific targets are clearly needed to help the nearly 35% of patients who still die from the disease. While RARA is a known druggable target in acute promyelocytic leukemia (APL), the utility of using retinoic acid agonists in non-APL AML has not proven consistently beneficial. Super enhancers (SEs), large regions of highly active chromatin, define cell state and cell identity by regulating oncogenes in many cancers. Recent enhancer profiling of 66 adult non-APL AML patient samples revealed SE-defined, prognostically relevant subgroups. An SE was detected at the retinoic acid receptor alpha (RARA) gene locus in 59% of the samples, which were sensitive to the second-generation retinoic acid agonist tamibarotene which has led to a phase II clinical trial. This study confirms that characterization of the chromatin-defined dependencies in specific cancers can pinpoint targets which can be drugged. We are delineating the transcriptional regulation of pediatric AML (pAML) by SE analysis, which has already elucidated deeper insights into pediatric leukemogenesis, as typified by strong RARA dependence in a majority of pAML.
Methods: Three AML cell lines and 19 pAML primary samples were enhancer profiled by H3K27Ac chromatin immunoprecipitation followed by massively parallel sequencing (ChIP-seq). SEs were detected and assigned to genes using the rank ordering of super-enhancers (ROSE) algorithm. Tamibarotene treatment of cell lines and patient samples were assessed for gene and protein expression changes and phenotypic differences. For in vivo assessment, 200,000 cells of a RARA SE+ pAML patient sample were injected into each NSGS mouse by tail-vein injection. One week after injection, tamibarotene (6mg/kg) or vehicle treatment was initiated by gavage (n=7 each arm). Peripheral blood monitoring of leukemia burden was determined flow cytometry.
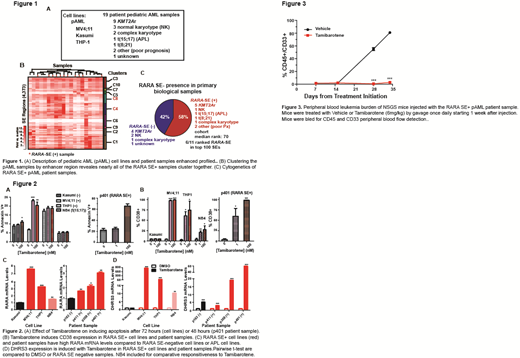
Results: The primary pAML sample cohort encompassed the diverse pAML cytogenetic subtypes (Fig 1a), with an overrepresentation of KMT2A rearrangements (n=9, 47%). The number of unique enhancer regions was nearly saturated in the 19 samples. Median SE size was 3,780bp, much larger than the 511bp of typical enhancers. When SE regions across all samples were clustered together, a RARA SE was seen in two of the ten clusters (Fig 1b). Eleven of the 19 samples (58%) contained a RARA SE, crossing multiple cytogenetic subtypes (Fig 1c). Tamibarotene treatment of RARA SE+ pAML cell lines and patient samples suppressed proliferation and increased apoptosis (detected by annexin V+), with minimal effect in Kasumi, a pAML cell line without a RARA SE (Fig 2a). In the RARA SE+ cell lines and samples only, tamibarotene increased CD38 (a myeloid differentiation marker usually suppressed by ligand-unbound RARA) (Fig 2b). High RARA mRNA levels confirmed the SE assignments in both cell lines and patient samples (Fig 2c). Tamibarotene induced the transcription of DHRS3 (another RARA target gene used as a pharmacodynamic biomarker in the adult tamibarotene phase II trial) (Fig 2d). Tamibarotene suppressed colony formation ability in RARA SE+ cell lines. An ongoing RARA SE+ patient-derived pAML xenograft confirmed tamibarotene markedly suppressed disease progression (Fig 3), with vehicle mice requiring euthanasia 42 days after tail-vein injection for significant disease burden, while the tamibarotene treated mice continued to be well-appearing at the same timepoint.
Conclusion: We have profiled the enhancer landscapes of 19 primary pAML samples, the largest dataset of its kind, and seen a high frequency of a RARA SE in pAML. Tamibarotene has anti-proliferative, proapoptotic, and on-target pro-differentiation effects in RARA SE+ pAML in vitro and marked anti-leukemia activity in vivo. Given these positive findings, we are evaluating combinations of other AML-active agents with tamibarotene. Additionally, as there is a range of sensitivity to tamibarotene in RARA SE+ samples, we are also interrogating other SE-regulated genes interacting with RARA that may predict degree of response or resistance to tamibarotene. Our studies confirm that studying the transcriptional regulation of pAML samples through SE analysis can identify druggable targets and also lay the preclinical foundation for a biomarker-defined tamibarotene trial in pediatric AML.
Wei:NHI NHLBI Grant: Other: received funding . Lin:Syros Pharmaceuticals: Equity Ownership, Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.