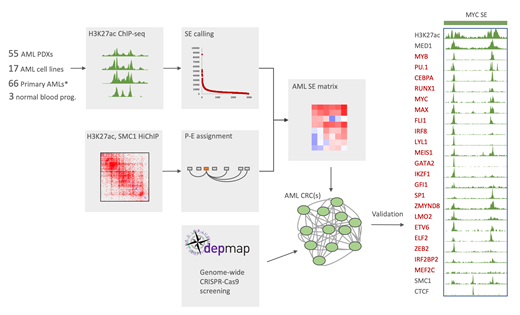
Core transcriptional regulatory circuitries (CRCs) are tightly integrated networks of master transcription factors (TFs) that establish and maintain lineage-specific programs of gene expression. We hypothesized that divergent CRCs establish distinct subtypes of acute myeloid leukemia (AML). CRCs are defined as sets of master TFs that are marked by superenhancers (SEs) and bind to their own genes and those of the other core TFs, forming feed-forward auto-regulatory loops. We have performed large-scale H3K27ac ChIP-seq experiments to map the SE landscape in 20 AML cell lines, 3 normal hematopoietic tissues and 50 patient-derived xenograft (PDX) models of human AML. These experiments highlighted a core set of SE-marked, highly expressed TF genes shared by all the examined AML subtypes, corresponding to a putative pan-AML CRC. Importantly, a significant majority (>70%) of these transcription factors correspond to AML-specific genetic dependencies in the Project Achilles genome-scale RNAi and CRISPR-Cas9 screening efforts undertaken by colleagues at the Broad Institute, confirming the specific reliance of AML on these TFs for survival. We reasoned that CRCs can be predicted by integrating the epigenomic and functional dependency datasets. Indeed, intersecting SE-marked TF genes with preferential AML dependencies resulted in 32 candidate TFs. We have validated these TFs as AML dependencies in a low-throughput system with lentiviral delivery of Cas9 and specific gRNAs. ChiP-seq experiments with antibodies against 23 of these candidates (GATA2, PU1, IRF8, GSE1, GFI1, MEIS1, LYL1, CEBPA, MEF2D, MEF2C, IKZF1, ZEB2, FLI1, ETV6, ELF2, MAX, RUNX1, MYB, IRF2BP2, LMO2, SP1, ZMYND8, MYC) resulted in nearly 100% validation rate. They demonstrated that CRC TFs tend to co-occupy DNA and bind their own and each other's promoters and SEs, suggesting that CRC members function in higher-order chromatin complexes and establish reciprocal feed-forward regulatory loops. Analysis of TF co-binding revealed 237,636 unique binding sites, with most occupied by at least two CRC TFs. Specific combinatorial patterns of TF binding appear to be associated with promoters, enhancers and super-enhancers. Importantly, in addition to the pan-AML CRCs, highly specific dependencies restricted only to a subset of AML cell lines can be accurately predicted from examination of divergent (subtype-specific) AML CRCs, lending support to our hypothesis that context-specific vulnerabilities can be robustly inferred from a systematic study of TF circuits. Specifically, we identified MEF2D and IRF8 as TFs that are selectively marked by SEs in a subset of AML cell lines and PDXs, most notably in samples carrying an MLL rearrangement. At the same time, these genes are strong preferential dependencies in a subset of AML cell lines, most of which also carry an MLL fusion. This suggests specific roles of IRF8 and MEF2D in MLL-induced leukemogenesis. Interestingly, functional dependency scores for these two TFs show an extremely high degree of correlation, indicating tightly integrated functions. While IRF8 is a known regulator of macrophage/dendritic cell function, MEF2D has no recognized roles in hematopoiesis. We have validated MEF2D as a dependency in a low-throughput CRISPR-Cas9 drop out experiment in an MLL-rearranged cell line. At the same time, we observed no functional effect of MEF2D knock out in a human CD34+ cell colony forming assay. This confirms context-specific transcriptional addiction to MEF2D induced by an MLL fusion and suggests a potential "Achilles heel" for leukemia-specific therapy with little or no detrimental effects on normal hematopoiesis. In summary, our data allow us to draw the following conclusions: 1) Intersection of lineage-restricted gene dependencies with SE profiling permits highly specific discovery of CRCs. 2) Transcriptional control in AML is orchestrated by a large CRC of >30 essential TFs. 3) Divergent CRCs are diagnostic of cancer- and context-specific transcriptional addiction. 4) AML CRC is a highly integrated network of co-binding TFs that orchestrate both promoter- and enhancer-centric regulation.
Lin:Syros Pharmaceuticals: Equity Ownership, Patents & Royalties. Stegmaier:Novartis: Research Funding; Rigel Pharmaceuticals: Consultancy. Orkin:Syros: Consultancy; Novartis: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal