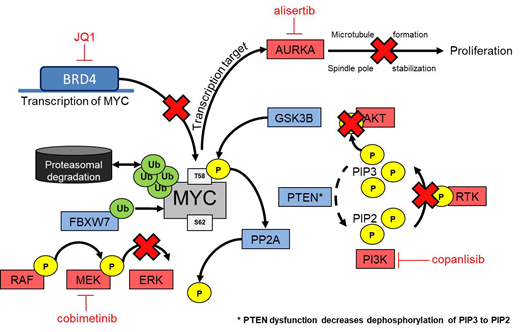
Therapy refractory and relapsed T-cell acute lymphoblastic leukemia (T-ALL) is still one of the most common causes for cancer related death in children. T-ALL is a highly heterogeneous disease with several established biomarkers, which promote its progression. These biomarkers are promising tools for patient stratification and can be used for targeted therapeutic approaches. For instance, PTEN is one of the most displayed mutations in pediatric T-ALL and plays an important role in proteolysis of the oncoprotein MYC. Therefore, it is not surprising that PTEN activity in T-ALL patients correlates with higher MYC protein levels. Importantly, PTEN mutations have recently been associated with poor overall survival in T-ALL patients, underscoring its importance for future treatment options using molecular targeted therapies.
In this study, we validated the importance of PTEN as a MYC regulator in T-ALL and elucidated PTEN mutational status as a stratification marker for T-ALL patients. Comparative quantification of PTEN, phosphorylated AKT and MYC in leukemic T-cell lines (n=10) was analyzed at protein level, relative to healthy T-cells. PTEN loss or deceased expression was observed in half of the screened leukemic T-cell lines with concomitant high MYC expression and increased AKT phosphorylation. To identify the drug sensitivity of PTEN mutant (PTENm) and PTEN wild type (PTENwt) cell lines, high-throughput drug screening was performed with 180 inhibitors, including among others kinase-, BET- and gamma secretase inhibitors, as well as conventional chemotherapeutics. PTENm cell lines showed remarkable sensitivity in nanomolar ranges against PI3K inhibitors compared to PTENwt (P=0.001). Anticipating that tumor cells eventually escape the killing effect of single pathway targeted drugs, the need for molecular identification and characterization of synergistic drug combinations is evident. Therefore, diverse compound combinations were tested in an 11 by 11 matrix of different concentrations in PTENm and PTENwt cell lines. Synergism was predicted with Combenefit software using Loewe model to assess putative additive versus synergistic effects. We observed that simultaneous use of copanlisib (PI3K inhibitor) with JQ1 (BET inhibitor) had a strong killing effect in PTENm cell lines. To assess target downregulation after 24 hours of drug treatment, lysates were taken and visualized using western blot. A strong decrease of MYC expression was observed already with a combination of low concentrations of copanlisib and JQ1 with subsequent higher apoptosis induction as measured by Annexin V/PI staining.
A second drug combination that showed synergy in PTENm was observed with copanlisib and alisertib, an Aurora kinase-A (AURKA) inhibitor, expecting changes in cell cycle and reduced MYC stability. Indeed, results of cell cycle analysis have revealed an inhibition during mitosis and cellular MYC levels were decreased, suggesting that this combination affects highly proliferative MYC-dependent tumor cells.
Contrary to PTENm, in PTENwt cell lines, PI3K inhibition alone showed no cytotoxic effect, but synergy was detected in combination with the MEK inhibitor cobimetinib. Moreover, in this synergistic combination MYC was effectively downregulated on protein level accompanied with changes in regulatory pathways including AKT and MEK. Interestingly, phosphorylation of MYC changed from the stabilizing site (S62) to the degradative site (T58), proposing the progress of MYC degradation.
In summary, we observed the importance of PTEN as a regulator for MYC proteolysis and related pathways including AKT and MEK. The loss of PTEN is accompanied with increased MYC. Synergy studies have observed effective drug combinations for PTENm and PTENwt with the aim to downregulate MYC and induce apoptosis in tumor cells. Based on these findings, PTEN-MYC axis seems suitable as a potential stratification marker for future therapy options in refractory and relapsed T-ALL. In the next step, promising drug combinations will be tested in a patient derived xenograft mouse model to validate these findings.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.