Introduction: ELN intermediate-risk AML poses considerable challenges to clinicians both in terms of accurate prognostication and optimal treatment. Indoleamine 2,3-dioxygenase 1 (IDO1) plays a central role as a mediator of immune tolerance in AML through the increase of Treg cells. IDO1 activity is negatively regulated by the BIN1 proto-oncogene. Herein, we analyzed the correlation between BIN1 and IDO1 expression in AML, also focusing on IDO1-interacting genes, with the aim to identify a predictive gene signature for OS.
Methods: Biological and clinical data of 732 patients with de novo AML were retrieved from public TCGA and HOVON datasets. Since details on chemotherapy regimens were not available in the HOVON dataset, we decided to exclude patients >= 65 years from survival analyses. IDO1-interacting genes were selected through a co-expression analysis performed on TCGA RNA-sequencing data accessed through cBioPortal. The best genes combination predicting overall survival was plotted in a gene expression score. Patients were split in three different groups using score quartiles as cut-off.
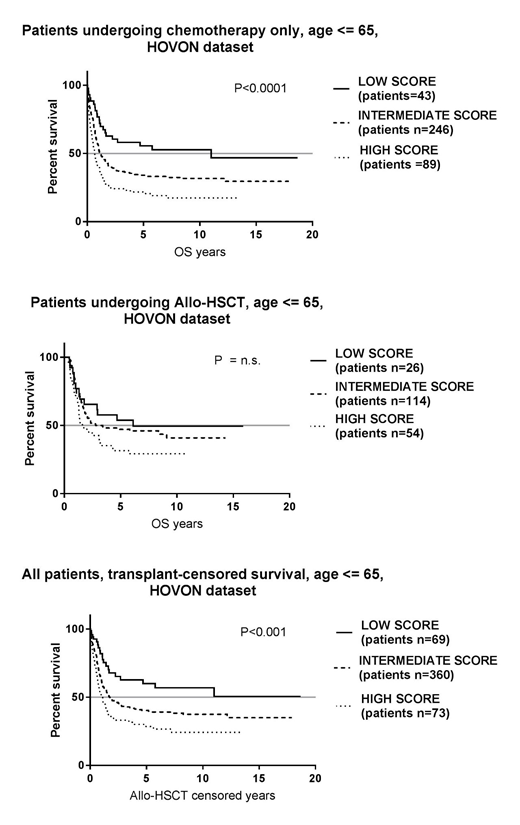
Results: In the HOVON dataset, IDO1 and BIN1 mRNA expression were negatively correlated (r = -0.40, P<0.0001). Our analysis of TCGA data identified PLXNC1 as an IDO1-interacting gene and a predictor of patient survival (median split of mRNA expression, P<0.001, survival analysis performed on the BloodSpot online portal). The correlation between PLXNC1 and IDO1 was validated in the HOVON dataset (r=-0.24, P<0.0001). PLXNC1 expression was combined with IDO1 and BIN1 expression to obtain the gene expression score. The 3-gene score predicted survival in ELN intermediate-risk patients who did not receive allogeneic HSCT both in the HOVON dataset (P<0.0001) and the TCGA dataset (P<0.05). In particular, the highest score values predicted the shortest OS.
Conclusions: Our study shows a negative correlation between IDO1 and BIN1 in AML, suggesting IDO1 inhibition by BIN1, and identifies for the first time PLXNC1, a receptor for semaphorines, as an IDO1-interacting gene potentially implicated in immune response regulation. This finding corroborates the role of IDO1 and its interacting genes in the promotion of a tolerogenic microenvironment in AML. Lastly, our gene expression score predicted OS in intermediate-risk AML patients not undergoing HSCT, a finding which has clinical implications for accurate patient stratification and for clinical decision making, i.e., bridging these patients to transplant.
Papayannidis:Pfizer: Honoraria; Amgen: Honoraria; Incyte: Honoraria; Novartis: Honoraria; Shire: Honoraria; Teva: Honoraria. Cavo:celgene: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: travel accommodations, Speakers Bureau; janssen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Other: travel accommodations, Speakers Bureau; bms: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; sanofi: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; novartis: Honoraria; takeda: Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; amgen: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Speakers Bureau; AbbVie: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees. Rutella:MacroGenics, Inc.: Research Funding; NanoString Technologies, Inc.: Research Funding; Kura Oncology: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.