Background
At diagnosis, recurrent genomic aberrations - aneuploidy and oncogene fusion that drive B-lymphoblastic leukemia - best predict treatment response. Currently, karyotyping and RT-PCR can discern only 50% of the genomic aberrations driving childhood B-ALL. RNA sequencing (RNA-Seq) interrogates the whole transcriptome through deep sequencing. Using RNA-Seq to study expressed SNP variants and expression level of each chromosome, we can identify their copy number; this enables digital karyotyping. For each patient, this digital karyotyping together with oncogene fusion sequencing and transcriptome profiling allow full molecular interrogation of the genomic aberrations that drive leukemia. We molecular subtyped and risk assigned 377 children with B-lymphoblastic leukemia using RNA-Seq. We showed that in childhood B-lymphoblastic leukemia, a single platform RNA-seq significantly improved risk assignment.
Methods
Whole transcriptome RNA-Seq was performed on the diagnostic bone marrow or peripheral blood samples of 377 children enrolled on the Ma-Spore ALL 2003 and Ma-Spore ALL 2010 studies. Oncogene fusions, variant calling, hierarchical clustering and gross chromosomal copy number calling were performed using RNA-Seq. These are combined for molecular assignment. As compared with conventional risk stratification using a combination of cytogenetics, DNA index and oncogene fusion panel, risk assignment including newly discovered molecular subtypes are made using RNA-Seq.
Results
Patients are classified into 20 molecularly distinct subtypes (Table 1) in 5 categories:
1. Oncogene fusion category with 10 subtypes.
2. Gene expression profiles with 5 subtypes.
3. Aneuploidy defines 3 subtypes, namely high hyperdiploidy, hypodiploidy and near haploid.
4. Biallelic mutation of PAX5 characterized by PAX5 P80R.
5. B-others.
The 20 molecular subtypes are then risk assigned to Low, Intermediate or High risk groups (Table 1). Specifically,
1. Low Risk (LR) - 66% (250/377) of the cohort
2. Intermediate Risk (IR) - 15% (57 of 377) of the cohort
3. High risk (HR) - 12% (44/377) of the cohort
4. B-others - 7% (26/377) of the cohort
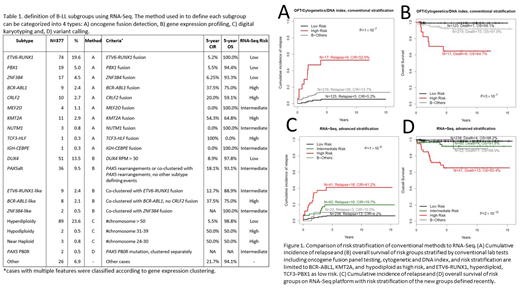
In the patients with available survival data, compared to 39% by conventional methods, RNA-Seq improves risk assignment by assigning 93% of the total cohort into a distinct risk group (Figure 1). The number of LR patients by RNA-Seq nearly doubled (RNA-Seq n=238 vs conventional n=125), with comparable cumulative incidence of relapse (CIR; 6.2% vs 5.2%) and overall survival (OS; 98.2% vs 99.1%). The number of HR patients also increased >2-fold (RNA-Seq n=41 vs conventional n=17), with a slightly lower CIR (41.2% vs 52.9%) and a similar OS (65.4% vs 64.7%). The RNA-Seq defined risk groups have more significantly distinct outcome (P-value comparing CIR: 1×10-8 vs1×10-7; P-value comparing OS: 2×10-12 vs3×10-7; Figure 1). In conventional risk assignment, 67% of the relapses and 68% of the deaths are in B-Others (61% of patients). Using RNA-Seq, HR (11% of patients) accounts for the most relapses (38%) and deaths (59%) while B-Others (7% of patients) account for 7% of relapses and 5% of deaths.
In multivariate analysis using competing risk regression on CIR together with age, gender, WBC and Day 33 MRD, only risk stratification by RNA-Seq and Day 33 MRD remained prognostically significant (RNA-Seq subtype P=6.4×10-4 or 5.9×10-3 for HR or IR vs SR; MRD P=1.3×10-5 or 0.020 for HR or IR vs SR). While in Cox proportional hazards regression on OS, Day 33 MRD lost its significance; RNA-Seq risk stratification is the most significant factor that predicts OS (RNA-Seq subtype P=3.4×10-4 or 0.27 for HR or IR vs SR; MRD P=0.19 or 0.24 for HR or IR vs SR).
Conclusion
When compared to conventional karyotyping and RT-PCR, in B-lymphoblastic leukemia, RNA-Seq significantly improved molecular diagnosis and risk assignment; it assigns 93% of patients into a distinct molecular and risk group. In Cox proportional hazards regression on OS, Day 33 MRD lost its prognostic significance, and RNA-Seq risk stratification assignment is the only significant predictive factor of overall survival.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.