In this issue of Blood, Li et al1 report age and co-mutation contexts wherein FLT3 internal tandem duplication (ITD) orchestrates unique transcriptional and epigenetic programs to deliver distinct functional outputs from myeloid progenitor cells.
Constitutive activating FLT3-ITD mutations are common in both adult and pediatric acute myeloid leukemia (AML). With the advent of FLT3-ITD directed tyrosine kinase inhibitors (TKI) over the past two decades the field has shifted its focus toward understanding FLT3-ITD signaling in the context of these FLT3-target inhibitors, with less effort aimed toward discovery and elucidation of FLT3-ITD leukemogenic mechanisms. Previous work demonstrated that knock-in of the human ITD mutation into the endogenous mouse Flt3 gene was not sufficient to produce AML.2,3 In patients with AML, regardless of age, FLT3-ITD mutations occur relatively late in AML pathogenesis and are observed in combination with a variety of mutational partners. Indeed, studies demonstrate that FLT3-ITD cooperates with DNMT3A, TET2, RUNX1, NPM1c, MLL-PTD, NUP98-HOXD13, and CBFB-MYH11 mutations to produce AML in mice.4-10 Despite the growing number of bona fide cooperating mutations leading to AML pathogenesis with FLT3-ITD, several questions remain. How this mutation contributes to profoundly different diseases in children and adults remains elusive. How FLT3-ITD signaling output changes as it is introduced with different co-occurring mutations remains vastly understudied. Whether the mutations that are coincident with FLT3-ITD, or the age of the patient, or both, are more important for specific disease characteristics, is still not clear. FLT3-ITD’s ability to enhance cell proliferation via STAT/extracellular signal-regulated kinase activation in leukemogenesis is well known, and this, coupled with continued improvements in potency and efficacy of FLT3 TKIs, has cast a shadow over identifying more intricate, context-specific functions of FLT3-ITD. Li et al took a completely new approach to understanding novel FLT3-ITD biology by comparing its role in models of pediatric vs adult AML, and in doing so, begin to answer the open questions lurking in the shadows.
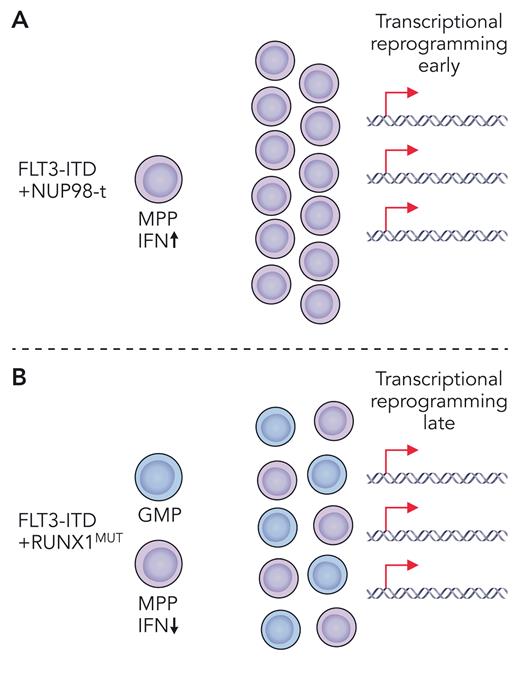
Driver gene alterations that co-occur with FLT3-ITD tend to be more age specific with chromosomal rearrangements, including NUP98-t and MLL-t, more common in pediatric AML, and single-gene mutations, such as DNMT3A and RUNX1 loss-of-function mutations, more common in adult AML. Based on coincidence in human AML, Li et al selected previously characterized mouse models of Flt3-ITD, harboring either NUP98-HOXD13 or RUNX1 deletion (RUNX1MUT) as representative of pediatric and adult coincident mutations, respectively. To level the playing field when making comparisons across these two different AML models, several key variables including the age of mice and the premalignant cell type were carefully controlled, and heterogeneity of the cell population was accounted for by single-cell RNA sequencing. This allowed direct comparison of Flt3-ITD’s contribution to the transcriptionally and immunophenotypically defined cell states in pediatric NUP98-HOXD13 vs adult RUNX1MUT contexts. In young mice, Li et al found that transcriptional reprogramming occurred earlier in Flt3-ITD multipotent progenitors (MPPs) coexpressing the fusion oncoprotein NUP98-HOXD13 (see figure panel A) and later in Flt3-ITD/RUNX1 co-mutant MPPs (see figure panel B). Despite both sets of mice having the same constitutively active Flt3-ITD mutation, very little overlap was observed in their leukemogenic gene expression profiles. These uniquely coordinated expression profiles also distinguished older mice with more advanced disease, as well as pediatric vs adult patients with AML. At the single-cell level, Flt3-ITD caused expansion of a granulocyte/monocyte progenitor (GMP)–like population and emergence of a new MPP-like population. The addition of RUNX1MUT co-mutation primarily enlarged these populations. Conversely, NUP98-HOXD13 drove development of a totally new and unique MPP-like population, which was also dominant with co-mutation of Flt3-ITD. Thus, Flt3-ITD/RUNX1MUT and Flt3-ITD/NUP98-HOXD13 each produce transcriptionally divergent cell states by distinct mechanisms (see figure). These novel insights demonstrate that FLT3-ITD is much more dynamic than previously thought. Moving forward, we should broaden how, and what, we consider as roles of FLT3-ITD in AML pathogenesis, keeping in mind that mutational and developmental contexts are key.
Depiction of two different mouse models of pediatric (A) and adult (B) Flt3-ITD co-mutant myeloid leukemia. (A) Flt3-ITD plus NUP98-translocation (NUP98-t) leads to early transcriptional reprogramming with unique upregulation of type 1 interferon. Co-mutant MPPs expand to give rise to AML. (B) Flt3-ITD plus RUNX1MUT leads to late transcriptional reprogramming and expansion of co-mutant MPPs and GMPs giving rise to AML. Professional illustration by Patrick Lane, ScEYEnce Studios.
Depiction of two different mouse models of pediatric (A) and adult (B) Flt3-ITD co-mutant myeloid leukemia. (A) Flt3-ITD plus NUP98-translocation (NUP98-t) leads to early transcriptional reprogramming with unique upregulation of type 1 interferon. Co-mutant MPPs expand to give rise to AML. (B) Flt3-ITD plus RUNX1MUT leads to late transcriptional reprogramming and expansion of co-mutant MPPs and GMPs giving rise to AML. Professional illustration by Patrick Lane, ScEYEnce Studios.
Critically, studies by Li et al identified a novel context-specific dependency of Flt3-ITD/NUP98-HOXD13 co-mutant MPPs on type 1 interferon (see figure panel A). Flt3-ITD/NUP98-HOXD13 but not Flt3-ITD/RUNX1 MPPs were sensitive to deletion of Ifnar1, resulting in significantly delayed leukemogenesis and prolonged survival. Future studies will need to understand whether type 1 interferon signaling can be leveraged therapeutically for treatment of pediatric FLT3-ITD mutant AML. In summary, the functional output of FLT3-ITD signaling is dynamic and has age-, cell-type–, and co-mutation-specific dependencies. This lesson in FLT3-ITD signaling will not soon be forgotten and will affect how we think about responsiveness to TKIs and other therapies in the future in different FLT3-ITD mutant AML subtypes.
Conflict-of-interest disclosure: S.E.M. has received research funding from CellCentric.