Clonal hematopoiesis of indeterminate potential (CHIP) is a common age-related condition associated with increased incidence of hematologic malignancies and overall mortality, but distinguishing true CHIP mutations from sequencing errors, rare germ line variants, and passenger mutations remains a major challenge for scientists, molecular pathologists, and practicing hematologists alike. In this issue of Blood, Vlasschaert et al1 report a novel method to increase the accuracy of true somatic variant calls in people with CHIP. By combining sequencing-based filtering and novel population-based filtering including age and TERT promoter polymorphism, the authors refined the list of likely true CHIP variants. Moreover, they demonstrated that this new approach resulted in a significant enrichment for variants that were strongly associated with clinical outcomes such as the incidence of myeloid malignancies and overall mortality.
Somatic mutations are an inevitable consequence of cell division. Every tissue accumulates mutations over time, resulting in the generation of cells with different genotypes, the phenomenon known as somatic mosaicism.2 Given their stochastic nature, the vast majority of somatic mutations are functionally inconsequential, but occasionally, they may result in increased hematopoietic stem cell fitness leading to relative clonal expansion, also known as CHIP.3 Not surprisingly, genes mutated in CHIP overlap with genes frequently mutated in myeloid malignancies. In addition to its malignant potential, CHIP has been associated with a plethora of nonmalignant, mostly inflammation-mediated conditions.4
Somatic CHIP mutations are detected by DNA sequencing. The distinction between functional somatic variants (driver mutations) and sequencing errors, rare germ line variants, and passenger mutations (nonfunctional mutations present in the expanded clone) remains problematic. Certain approaches such as polymerase chain reaction (PCR)-free sequencing may minimize the rate of sequencing errors, and concurrent sequencing of matched tumor and germ line tissues may help differentiate between somatic and germ line variants. Unfortunately, these strategies cannot be applied retrospectively to large cohorts of whole genome or whole exome sequencing. Yet the majority of associations between CHIP and various diseases have been described using these invaluable resources. Can a novel scheme be applied to refine the list of true CHIP variants?
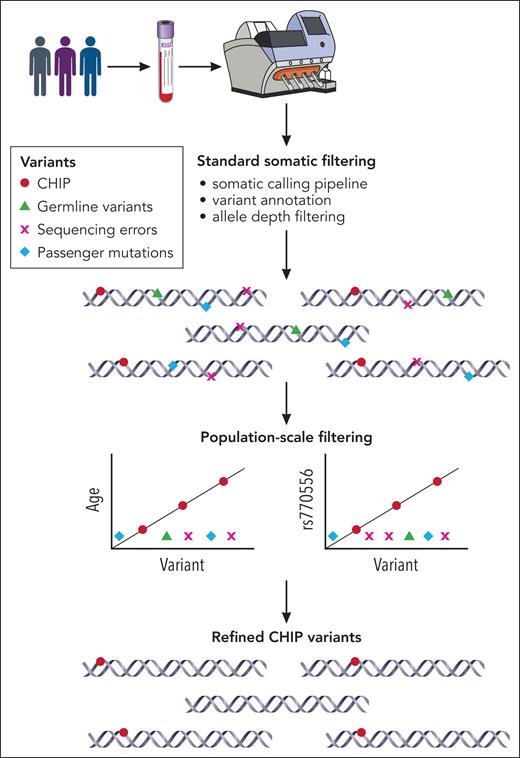
To address this important problem, Vlasschaert et al used a combination of sequencing metrics, variant annotation, and a novel population-based association filtering to further improve and standardize the bioinformatics method for CHIP calling. The authors analyzed sequencing data from over 550 000 individuals from the UK Biobank complete whole exome cohort and the All of Us Research Program initial whole genome cohort. Age and single nucleotide polymorphism in TERT promoter (rs7705526) have been strongly associated with CHIP.5,6 The authors leveraged this association to exclude true CHIP from the sequencing artifacts (see figure). This was particularly useful for ASXL1 variants. Somatic mutations in ASXL1 are common in CHIP and hematologic malignancies. Some of the mutations fall within the 8 monoguanine homopolymer sequence and may represent a PCR amplification error. For example, previously described mutation p.G646Wfs∗12 has been reported as a true somatic variant but also as a sequencing artifact, particularly when present at low variant allele frequency (VAF).7 The authors demonstrated that ASXL1 p.G646Wfs∗12 mutation with VAF >10% was indeed a true somatic mutation associated with age and an 18-fold increased risk of myeloid cancer.
A stepwise approach to CHIP variant identification. The multistep process of CHIP variant filtering using standard somatic filtering and novel population-based filter. Because CHIP is strongly associated with age and TERT promoter polymorphism (rs770556), most sequencing errors, germ line variants, and passenger mutations are excluded as they lack the associations with age and TERT polymorphism. Professional illustration by Patrick Lane, ScEYEnce Studios.
A stepwise approach to CHIP variant identification. The multistep process of CHIP variant filtering using standard somatic filtering and novel population-based filter. Because CHIP is strongly associated with age and TERT promoter polymorphism (rs770556), most sequencing errors, germ line variants, and passenger mutations are excluded as they lack the associations with age and TERT polymorphism. Professional illustration by Patrick Lane, ScEYEnce Studios.
Standard somatic variant filtering strategies excluded variants with VAF close to 50% as potential heterozygous germ line polymorphisms. However, some of these presumed germ line polymorphisms may represent large CHIP clones. This is of utmost clinical importance as patients with large CHIP are at the highest risk for leukemic transformation. Hence, the distinction between CHIP and germ line polymorphism may significantly influence not only surveillance strategies but also therapeutic decisions, particularly in patients with concurrent cytopenias.8 By examining whether the addition of an ostensible germ line variant improved or weakened the association with age, the authors defined the list of likely CHIP and likely germ line polymorphisms in TET2, DNMT3A, RUNX1, and TP53.
Another challenge in CHIP variant calling is to determine the minimum VAF threshold. This can be represented as the absolute number of reads supporting the variant, also known as minimum allele depth (minAD). It is now well recognized that small hematopoietic clones (VAF <1%) are present in nearly all individuals but rarely expand over time, and hence their functional and clinical significance is unclear.9,10 To identify the meaningful minAD, the authors tested the associations between putative variants in different minAD levels and age and the TERT promoter variant. They established that minAD of ≥5 was optimal for UK Biobank whole exome sequencing. This threshold demonstrated a strong association with increased mortality and the incidence of myeloid cancer reported in previous studies.
CHIP is a very dynamic area of research, not only within hematology as a premalignant condition but spanning across multiple disciplines and organ systems. Despite the remarkable progress made over the past decade in defining the biological and clinical consequences of CHIP, the consensus for CHIP calling has not been established. The article by Vlasschaert et al offers a practical approach to the CHIP variant definition and the first step in the standardization of our analytical approach. Further refinement of the sequencing and analytical process to increase reproducibility and scientific rigor will provide much-needed guidance for the molecular laboratories for CHIP variant reporting and for practicing clinicians to increase their confidence in the interpretation of molecular results.
Conflict-of-interest disclosure: The author declares no competing financial interests.