TO THE EDITOR:
Fibrin-associated large B-cell lymphoma (FA-LBCL) is a microscopic proliferation of large B cells arising in association with fibrinous debris in specific anatomic locations, including cardiac valves and myxomas, preexisting cysts or pseudocysts, and chronic hematomas or thrombi.1,2 Some of these locations are associated with the presence of foreign bodies, especially artificial cardiac valves and endovascular grafts, whereas others arise in the context of native tissue.
The pathophysiology of FA-LBCL is largely unknown; however, a few potential pathogenic mechanisms have been proposed. The lymphoma cells in most cases are positive for Epstein-Barr virus (EBV) and express EBV latent membrane proteins and EBV nuclear antigens (type III latency), which is unusual for lymphoma in immunocompetent patients because of the immunogenicity of these viral proteins. This has led to the hypothesis that the lymphoma cells are obliged to stay in protected anatomic spaces to evade immune surveillance. It has also been proposed that myxoma-associated cases of FA-LBCL may be protected from immune surveillance by the production of interleukin-6 by the myxoma3 and that upregulation of PD-L1 may also support immune evasion in some cases.2
Genetic characterization of FA-LBCL has been limited so far. Most of the reported cases that have been tested are negative for rearrangements of BCL2, BCL6, and MYC with the exception of 1 case with BCL6 rearrangement4 and 1 with MYC rearrangement.5 One case was characterized by array comparative genomic hybridization and showed no significant genomic losses or gains.2 In this study, we sought to understand the genomic landscape of this rare entity.
The case files of the pathology departments at the University of Michigan and City of Hope National Medical Center were reviewed, identifying 6 cases of FA-LBCL with archived formalin-fixed paraffin-embedded tissue sufficient for sequencing studies. DNA was extracted, and library preparation with whole-exome sequencing was performed using Illumina NovaSeq 6000 for cases 1 to 5. A targeted sequencing panel was performed for case 6.6 LymphGen7 was used for assignment. Immunohistochemistry (IHC) staining for PTEN, CA9, pSTAT3, and pAKT-T308 and in situ hybridization for EBV-encoded small RNA (EBER) was performed (supplemental Material, available on the Blood website). This research was approved by the institutional review boards of the University of Michigan and City of Hope and was conducted according to the Declaration of Helsinki.
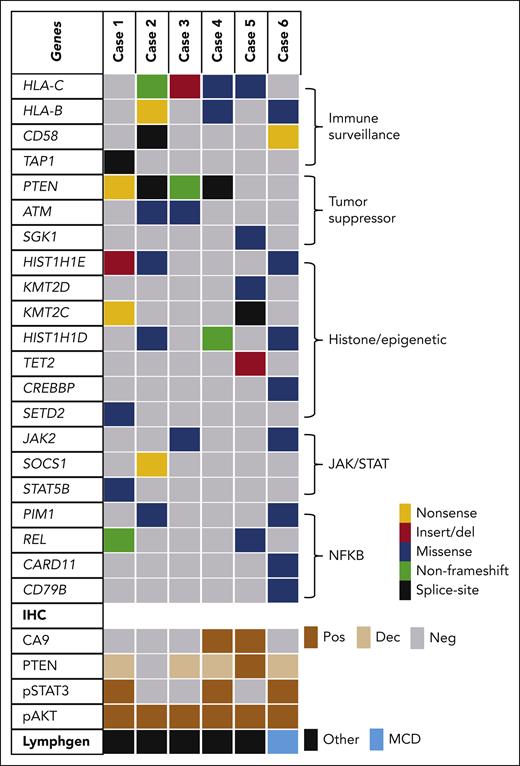
Mutation analysis was performed on 6 cases of FA-LBCL. The patients included 5 males and 1 female, ranging in age from 36 to 77 years (Table 1). Clinicopathologic features of 4 of these cases were previously reported prior to the availability of sequencing data.2,5,8 Five of the cases were positive for EBER, and case 6 was negative for EBER but had otherwise typical histopathologic and clinical features of FA-LBCL.8 The results of sequencing (Figure 1 and supplemental Table 2) showed frequent mutations in immune surveillance pathways, histone and chromatin modification genes (especially HIST1H1D and HIST1H1E), the tumor suppressor PTEN, and components of the JAK/STAT pathway.
Summary of genomic sequencing of FA-LBCL. Cases of FA-LBCL showed frequent mutations with immune surveillance (HLA-B, HLA-C, CD58), histone modification genes, JAK/STAT pathway, NF-κB, and tumor suppressor genes, particularly PTEN. Dec, decreased; MCD, MYD88 and CD79B group; Neg, negative; Pos, positive.
Summary of genomic sequencing of FA-LBCL. Cases of FA-LBCL showed frequent mutations with immune surveillance (HLA-B, HLA-C, CD58), histone modification genes, JAK/STAT pathway, NF-κB, and tumor suppressor genes, particularly PTEN. Dec, decreased; MCD, MYD88 and CD79B group; Neg, negative; Pos, positive.
All 6 cases had 1 or more mutation in genes encoding components of immune surveillance pathways including the class I major histocompatibility complex (MHC) components HLA-B and HLA-C, TAP1 (a transporter involved in class I MHC function), and CD58 (an adhesion molecule involved in T-cell activation). No mutations in B2M were detected. Frequent mutations in components of the class I MHC pathway have been reported in EBV+ diffuse large B-cell lymphoma (DLBCL) and EBV+ extranodal natural killer/T-cell lymphoma,9,10 suggesting that these mutations may be important for EBV immune evasion. Another similarity between EBV+ FA-LBCL (cases 1-5) and previously described cases of EBV+ DLBCL is the lack of MYD88 and CD79B mutations.9 Case 6 was negative for EBV based on EBER in situ hybridization and had mutations in CD79B p.Y196S and CARD11 p.K215T that have been previously shown to increase NF-κB pathway activation.11,12 EBV-encoded latent membrane proteins can also activate the NF-κB pathway, which might explain the paucity of NF-κB–activating mutations in EBV+ lymphomas.13
Four of 6 cases had mutations in the tumor-suppressor PTEN. Immunostaining showed decreased PTEN expression (supplemental Figures 1-4) in all cases except for case 5, which did not have a PTEN mutation. Case 5 had a mutation in SGK1 p.N139Y, which is also involved in the PI3K pathway. All 6 cases were positive for phosphorylation of AKT at the T308 residue, based on phospho-specific IHC staining (supplemental Figures 1 and 2). AKT-T308 is preferentially phosphorylated by PI3K; therefore, activation of the PI3K pathway appears to be a common feature of FA-LBCL. PTEN inactivation likely contributes to PI3K activation in some cases.
The pattern of tumor-suppressor inactivation in these cases of FA-LBCL was surprising because PTEN mutations14 and deletions15 occur most frequently in germinal center B-cell–type DLBCL,14 but FA-LBCL usually has a non–germinal center B-cell immunophenotype.2 In contrast, TP53 mutations are relatively frequent in EBV+ DLBCL9 and pyothorax-associated lymphoma16 but were not detected in these 6 cases of FA-LBCL. The paucity of TP53 mutations in FA-LBCL may contribute to its relatively indolent behavior compared with EBV+ DLBCL and pyothorax-associated lymphoma.
Four of 6 cases had mutations in the JAK/STAT signaling pathway, including JAK2, SOCS1, and STAT5B. IHC for phosphorylated STAT3 (pSTAT3) was performed to evaluate for evidence of JAK/STAT activation, and cases 1, 4, and 6 were positive for pSTAT3. Case 6 had an activating mutation in the protein kinase domain of JAK2 (p.N1108S)17; however, the mechanism for STAT3 activation in cases 1 and 4 was unclear. Case 2 had a nonsense mutation of SOCS1 (p.C42∗) at a high variant allele frequency (69%). This mutation should result in inactivation of SOCS1, which is a negative regulator of JAK/STAT signaling; however, pSTAT3 IHC staining result was surprisingly negative in this case.
The presence of JAK/STAT pathway mutations has been described previously in breast-implant–associated anaplastic large cell lymphoma (BIA-ALCL) and BIA-DLBCL.18,19 These entities have similar features to FA-LBCL, including association with cavitary spaces, frequent incidental and/or microscopic presentation, and relatively indolent behavior. Recurrent JAK/STAT mutations may represent another common feature among these entities. However, JAK/STAT pathway activation does not appear to be as consistent of a feature in FA-LBCL compared with BIA-ALCL. All described cases of BIA-ALCL have been positive for pSTAT3 IHC regardless of mutational status,18 but only 3 of 6 cases of FA-LBCL in the current study were positive for pSTAT3 IHC. Similar to BIA-ALCL,20 2 of 6 cases showed strong expression of CA9 (carbonic anhydrase-9), which is upregulated in hypoxic states. This result indicates that some cases of FA-LBCL have hypoxia-associated proteins.
Frequent PTEN mutations in FA-LBCL and positivity for pAKT-T308 in all 6 cases suggest that PI3K/AKT pathway inhibition could be a therapeutic option for FA-LBCL when the lesional tissue cannot be completely excised. For example, a previous study found that a small molecule inhibitor of AKT selectively induced apoptosis in PTEN-deficient DLBCL cell lines.21 JAK/STAT inhibition could also be a therapeutic option to consider for FA-LBCL. This cohort of 6 cases included several anatomic sites of involvement; however, there were too few cases to evaluate whether any of the genetic abnormalities were associated with specific anatomic sites. After more cases have been characterized, it will be interesting to see whether there are any significant differences in the genetic profile of FA-LBCL associated with distinctive sites, such as cardiac myxomas or endovascular grafts.
Acknowledgments
Research reported in this letter included work performed in the City of Hope Pathology Core and Integrated Genomics Core supported by the National Cancer Institute, National Institutes of Health (grant P30CA033572) and the City of Hope Lymphoma SPORE (grant P50CA107399).
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Authorship
Contribution: D.F.B., A.P., and J.Y.S. conceived and designed the study; D.F.B., A.P., E.W., J.H., A.L., R.J., L.S., W.Z., and J.Y.S. collected and assembled the data; W.Z., D.F.B., R.J., A.P., and J.Y.S. performed data analysis; D.F.B. prepared the first draft; and all authors helped with revisions.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Daniel F. Boyer, Department of Pathology, University of Michigan, NCRC Bldg 36, 1361-07, Ann Arbor, MI 48109; e-mail: dfboyer@med.umich.edu.
References
Author notes
The dataset has been uploaded to the National Center for Biotechnology Information BioProject database with the public release date set for 23 August 2023. BioProject ID: PRJNA943222. The data are also included in this article as supplemental Table 2.
The online version of this article contains a data supplement.