The paracaspase mucosa-associated lymphoid tissue 1 (MALT1) is a protease and scaffold protein, essential in propagating signals from the B-cell receptor (BCR) and various oncogenic proteins to NF-κB; hence, it is a potential therapeutic target for treatment of a variety of B-cell malignancies. In this issue of Blood, Wimberger et al study the protease function of MALT1 in chronic BCR signaling and signaling by oncogenic CARD11 in activated B cell–like diffuse large B-cell lymphoma (ABC-DLBCL) and of the API2-MALT1 fusion oncoprotein in MALT lymphoma.1
Most mature B-cell malignancies depend on (chronic) BCR- and/or oncogenic protein-propagated constitutive canonical NF-κB signaling for their growth and survival. This is illustrated by recurrent gain-of-function mutations in CD79, CARD11, BCL10, and MYD88 and loss-of-function deletions/mutations in TNFAIP3 (A20) in DLBCL; the chromosomal translocation product API2-MALT1 in MALT lymphoma; and the clinical efficacy of the Bruton tyrosine kinase (BTK) inhibitor ibrutinib in chronic lymphocytic leukemia (CLL), mantle cell lymphoma (MCL), marginal zone lymphoma, and DLBCL.
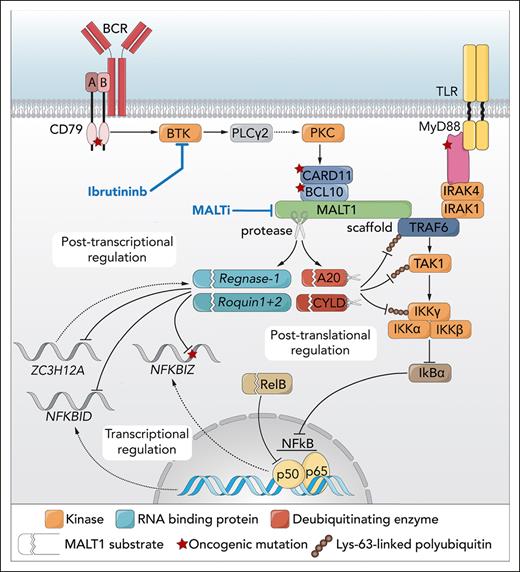
Formation of a complex composed of the caspase recruitment domain family member 11 (CARD11), B-cell lymphoma 10 (BCL10), and MALT1, the CBM complex, is a key intermediate in signaling of the BCR to canonical NF-κB activation.2 MALT1 functions as a scaffold protein, allowing recruitment and activation of the E3-ubiquitin ligase tumor necrosis factor receptor-associated factor 6 (TRAF6). TRAF6 mediates Lys-63–linked polyubiquitination of the regulatory γ subunit of the IκB kinase (IKK) complex (IKK-γ or NEMO), thereby contributing to its activation, which results in phosphorylation and proteasomal degradation of IκB and dissociation and nuclear translocation of NF-κB (see figure).2,3 Next to its function as a scaffold protein, MALT1 possesses protease activity and is able to cleave and inactivate negative regulators of canonical NF-κB signaling, such as A20, RelB, and CYLD (see figure).1-3 Recently, it was demonstrated that low expression of the deubiquitinating enzyme CYLD, which can hydrolyze Lys-63–linked ubiquitin chains of TRAF6 and IKK-γ, is associated with a poor prognosis of patients with ABC-DLBCL and MCL and that chronic BCR signaling propagates MALT1-mediated cleavage and proteasomal degradation of CYLD, thereby contributing to canonical NF-κB signaling and growth of BCR-dependent lymphomas.3
Simplified model of the role of MALT1 and regnase-1 in B-cell lymphoma. BCR signaling through BTK mediates the formation of a complex of CARD11, BCL10, and MALT1. Oligomerized MALT1, but also the oncoprotein API2-MALT1, functions as a scaffold protein, binding the E3 ubiquitin ligase TRAF6. In parallel, Toll-like receptor (TLR) signaling may result in MyD88-dependent recruitment of IRAK4 and IRAK1, which can also associate with TRAF6. TRAF6 promotes Lys-63–linked ubiquitination of TRAF6 itself as well as TAK1 and IKK-γ/NEMO, resulting in their interaction and TAK1-mediated activation of IKK. IKK mediates phosphorylation and degradation of IκBα, allowing nuclear translocation of NF-κB dimers. In addition, MALT1 (and API2-MALT1) is a protease that cleaves various negative regulators of NF-κB, such as A20, CYLD, and RelB. The deubiquitinating enzymes A20 and CYLD hydrolyze Lys-63–linked polyubiquitin chains of TRAF6, TAK1, and/or NEMO/IKK-γ, whereas RelB can interact with and repress activity of canonical NF-κB subunits; hence, MALT1-dependent inactivation of A20, CYLD, and RelB promotes NF-κB activation and cell growth. The study by Wimberger et al shows that MALT1 protease activity can also regulate gene expression independent of TRAF6/NF-κB, at the posttranscriptional level, by inactivation of the RNA-binding proteins regnase-1 and roquin-1/2, thereby promoting the stability of the NFKBIZ (IκBζ), NFKBID (IκBNS), and ZC3H12A (regnase-1) transcripts. Similar findings are presented for the API2-MALT1 oncoprotein. Professional illustration by Somersault18:24.
Simplified model of the role of MALT1 and regnase-1 in B-cell lymphoma. BCR signaling through BTK mediates the formation of a complex of CARD11, BCL10, and MALT1. Oligomerized MALT1, but also the oncoprotein API2-MALT1, functions as a scaffold protein, binding the E3 ubiquitin ligase TRAF6. In parallel, Toll-like receptor (TLR) signaling may result in MyD88-dependent recruitment of IRAK4 and IRAK1, which can also associate with TRAF6. TRAF6 promotes Lys-63–linked ubiquitination of TRAF6 itself as well as TAK1 and IKK-γ/NEMO, resulting in their interaction and TAK1-mediated activation of IKK. IKK mediates phosphorylation and degradation of IκBα, allowing nuclear translocation of NF-κB dimers. In addition, MALT1 (and API2-MALT1) is a protease that cleaves various negative regulators of NF-κB, such as A20, CYLD, and RelB. The deubiquitinating enzymes A20 and CYLD hydrolyze Lys-63–linked polyubiquitin chains of TRAF6, TAK1, and/or NEMO/IKK-γ, whereas RelB can interact with and repress activity of canonical NF-κB subunits; hence, MALT1-dependent inactivation of A20, CYLD, and RelB promotes NF-κB activation and cell growth. The study by Wimberger et al shows that MALT1 protease activity can also regulate gene expression independent of TRAF6/NF-κB, at the posttranscriptional level, by inactivation of the RNA-binding proteins regnase-1 and roquin-1/2, thereby promoting the stability of the NFKBIZ (IκBζ), NFKBID (IκBNS), and ZC3H12A (regnase-1) transcripts. Similar findings are presented for the API2-MALT1 oncoprotein. Professional illustration by Somersault18:24.
Underlining the critical role of MALT1 in propagating NF-κB activity, inhibition of MALT1 proteolytic activity impairs growth and survival of MCL and ABC-DLBCL in the preclinical setting, and various MALT1 inhibitors are currently under clinical investigation for mature B-cell malignancies (outlined below).2,3 However, despite these recent preclinical and clinical developments, as of yet the role of the MALT1 protease activity in lymphomagenesis remains incompletely understood.
In the current study, Wimberger et al combined CRISPR/Cas9-mediated knockout of MALT1 or TRAF6, ectopic expression of function-compromised MALT1 mutants, and pharmacological MALT1 inhibition to study canonical NF-κB signaling evoked by PMA/ionomycin stimulation (to mimic BCR signaling) and oncogenic CARD11. They demonstrate that most target genes are codependent on MALT1 protease activity and TRAF6 binding, reflecting the scaffold function of MALT1, for optimal expression. However, some genes are only MALT1 protease dependent. These included NFKBIZ (IκBζ), NFKBID (IκBNS), and ZC3H12A (regnase-1) (see figure), which are also highly expressed in ABC-DLBCL. Interestingly, of these genes, only NFKBIZ is under transcriptional control of NF-κB,1,4 indicating that the MALT1 protease exerts effects beyond NF-κB to control expression of IκBNS and regnase-1.
At the protein level, the RNA-binding protein (RBP) regnase-1 and other RBPs named roquin-1 and -2 were found to be constitutively cleaved and inactivated by MALT1,5,6 which was also observed in ABC-DLBCL cells (see figure). The cleavage of these RBPs, and the expression of the MALT1 protease-dependent genes, is repressed by treatment with the BTK inhibitor ibrutinib and MALT1 protease inhibitors. Furthermore, in ABC-DLBCL cells, the stability of the NFKBIZ and NFKBID transcripts is repressed by binding of the RBPs to their 3’UTRs, which is promoted by ibrutinib and MALT1 inhibitor treatment. This repression by the RBPs is abolished by previously identified7 recurrent mutations in the RBP binding sequence in the 3’UTR of NFKBIZ in ABC-DLBCL (see figure). Finally, similar results are presented for the protease activity of the API2-MALT1 oncoprotein, at the transcriptional, posttranscriptional, and posttranslational levels.
Taken together, these novel findings indicate that MALT1 protease activity controls gene expression not only at the transcriptional but also at the posttranscriptional level by cleaving RBPs, resulting in enhanced expression of NF-κB–dependent and –independent genes in ABC-DLBCL and MALT lymphoma.
This study provides novel insights into the molecular mechanisms underlying MALT1-mediated NF-κB signaling and how direct inhibition of MALT1 protease activity, but also indirect inhibition of MALT1 by other BCR signalosome inhibitors such as ibrutinib, may contribute to the antilymphoma effects of these drugs. These findings have clinical relevance for the development of therapeutic strategies aimed at targeting MALT1 or downstream effectors as treatment for (mature) B-cell malignancies. Furthermore, the identified MALT1 protease-specific target genes may serve as suitable biomarkers for clinical studies.
Despite promising preclinical results, the MALT1 inhibitory peptide z-VRPR-fmk and the irreversibly binding pharmacological inhibitor MI-2 turned out to be unsuitable for clinical use, whereas the reversible MALT1 inhibitory phenothiazine derivatives and the pyrazolopyrimidine derivatives (MLT-XXX) have not yet been studied clinically. However, for several other MALT1 (protease) inhibitors, ie, safimaltib (JNJ-67856633) (ClinicalTrials.gov: NCT03900598 and NCT04876092), ONO-7018 (CTX-177) (NCT05515406), SGR-1505 (NCT05544019), and ABBV-525 (NCT05618028), clinical trials are currently active for various (mature) B-cell malignancies. Of note, MALT1 inhibitors also have clinical potential for patients with primary resistance to ibrutinib, eg, patients with DLBCL with CARD11 or BCL10 mutations, or patients who developed secondary ibrutinib resistance, eg, patients with MCL or CLL with acquired mutations of the ibrutinib-binding cysteine residue in BTK.8,9
The novel insights provided by Wimberger et al and other recent studies,3,8-10 as well as future studies on the specific roles of the distinct protease and scaffold functions of MALT1, may pave the way toward precision treatment by informed combinations of small-molecule inhibitors or PROTAC degraders of specific kinases, ubiquitinating enzymes, or transcription factors downstream of MALT1, targeting, for example, TRAF6, IKK, or individual NF-κB subunits. Such an informed precision approach to selectively suppress (a combination of) distinct canonical or noncanonical NF-κB signals would provide a promising therapeutic strategy for mature B-cell malignancies.
Conflict-of-interest disclosure: The author declares no competing financial interests.