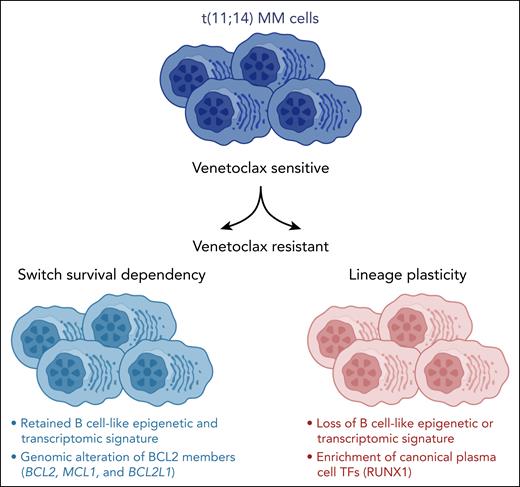
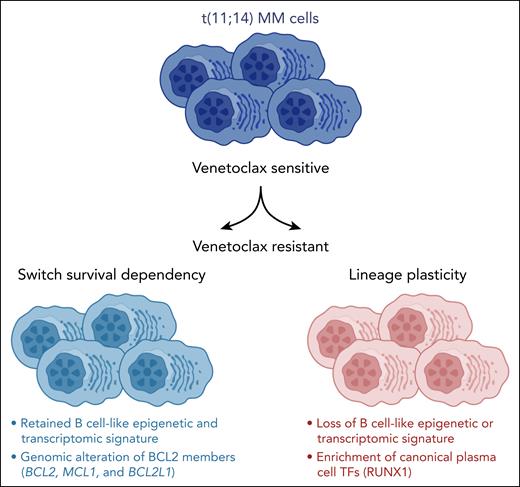
By applying single-cell epigenetics and transcriptomics, Leblay et al1 reveal in this issue of Blood that cellular lineage plasticity, due to a switch from a B-cell–like phenotype to a more canonical plasma cell–like state, contributes to drug-acquired resistance in patients with t(11;14)-positive multiple myeloma (MM) receiving venetoclax.
Venetoclax is an oral BH3 mimetic that effectively displaces proapoptotic proteins from the inhibitor of apoptosis B-cell lymphoma 2 (BCL-2). This compound has become standard of care, as a single agent and in combinations, in chronic lymphocytic leukemia (CLL) and, more recently, in acute myeloid leukemia. In MM, a phase 1 study showed promising efficacy as monotherapy in patients harboring a t(11;14) translocation, whereas initial results from the combination of venetoclax with the proteasome inhibitor, bortezomib, initially raised hope for its efficacy in non-t(11;14) patients.2,3 The phase 3 placebo-controlled BELLINI study did not show superior outcomes for this combination compared with bortezomib and dexamethasone, mainly due to higher mortality in patients receiving venetoclax. However, subgroup analyses revealed higher response rates and longer progression-free survival for t(11;14) patients and those with high expression of BCL-2.4 The first results of the ongoing registration phase 3 CANOVA trial of either venetoclax or pomalidomide in combination with dexamethasone in patients with a t(11;14) translocation are expected later this year.
Biomarker analyses from trials of venetoclax in MM2-4 and in vitro data have suggested that high BCL-2 expression, both in absolute terms and relative to the other antiapoptotic family members, Bcl-XL and myeloid cell leukemia (MCL)-1, as well as a more B-cell–like MM cell phenotype, may be more precise markers of venetoclax sensitivity than a t(11;14) translocation.5 Apart from inhibitors of the raf murine sarcoma viral oncogene homolog B (BRAF)/MEK/extracellular signal-regulated kinase pathway in patients with an activating BRAF V600E mutation, venetoclax is the only molecularly guided personalized treatment option currently available in MM.6
In the present study, Leblay et al explored the sensitivity of t(11;14) MM cells to venetoclax and mechanisms of acquired resistance. By integrating single-cell RNA and assay for transposase-accessible chromatin (ATAC) sequencing of patient samples, they identified the epigenetic and transcriptomic characteristics of these cells and found a B-cell–like signature enriched in t(11;14) MM cells.
More important, when comparing samples from patients before and after treatment with venetoclax, a shift toward a canonical plasma cell–like regulatory program was observed with partial or even complete loss of the B-cell–like phenotype in patients with acquired resistance to venetoclax, reminiscent of treatment-induced transcriptional lineage plasticity described in other cancers.7 However, MM is biologically highly complex, and each patient’s disease contains of a range of subclonal cell clusters.8 Accordingly, in some patients and, even then only in some of their subclonal clusters, the B-cell–like signatures were retained after treatment with venetoclax. In these patients, alternative mechanisms of resistance, such as upregulation of RUNX1, BCL2L1, or Mcl-1 by various mechanisms, including copy number changes, were observed, as recently described in CLL (see figure).9
In patients with t(11;14) myeloma, a loss of a “B-cell–like” epigenetic and transcriptomic signature with a gain of canonical plasma cell transcription factors (TFs) is observed at the time of resistance to venetoclax. MCL1 and BCL2L1 copy number gains and structural rearrangements are also linked to venetoclax resistance. See the visual abstract in the online version of the article by Leblay et al that begins on page 42.
In patients with t(11;14) myeloma, a loss of a “B-cell–like” epigenetic and transcriptomic signature with a gain of canonical plasma cell transcription factors (TFs) is observed at the time of resistance to venetoclax. MCL1 and BCL2L1 copy number gains and structural rearrangements are also linked to venetoclax resistance. See the visual abstract in the online version of the article by Leblay et al that begins on page 42.
Applying longitudinal single-cell interrogation of the epigenome, transcriptome, and genome, as reported here and recently by Poos et al,10 sheds light on the complex layers of cellular adaptations to standard and novel therapeutic options in MM. This approach will likely be useful for exploring responses and resistance for the emerging T-cell–engaging approaches. It is tempting to speculate that targeting regulome-driven cellular plasticity might represent a strategy to overcome treatment resistance and improve patient outcomes and is worthy of further investigation.
Conflict-of-interest disclosure: The author declares no competing financial interests.