Visual Abstract
Peripheral T-cell lymphomas (PTCLs) encompass a heterogeneous group of postthymic T-cell lymphomas with >30 distinct subtypes associated with varied clinicopathological features. Unfortunately, the overall survival of the major PTCL subtypes is dismal and has not improved for decades; thus, there is an urgent unmet clinical need to improve diagnosis, therapies, and clinical outcomes. The diagnosis is often challenging, requiring a combinatorial evaluation of clinical, morphologic, and immunophenotypic features. PTCL pathobiology is difficult to investigate due to enormous intertumor and intratumor heterogeneity, limited tissue availability, and the paucity of authentic T-cell lymphoma cell lines or genetically faithful animal models. The application of transcriptomic profiling and genomic sequencing has markedly accelerated the discovery of new biomarkers, molecular signatures, and genetic lesions, and some of the discoveries have been included in the revised World Health Organization or International Consensus Classification. Genome-wide investigations have revealed the mutational landscape and transcriptomic profiles of PTCL entities, defined the cell of origin as a major determinant of T-cell lymphoma biology, and allowed for the refinement of biologically and clinically meaningful entities for precision therapy. In this review, we prioritize the discussion on common nodal PTCL subtypes together with 2 virus-associated T-cell and natural killer cell lymphomas. We succinctly review normal T-cell development, differentiation, and T-cell receptor signaling as they relate to PTCL pathogenesis and biology. This review will facilitate a better biological understanding of the different PTCL entities and their stratification for additional studies and target-directed clinical trials.
Introduction
Peripheral T-cell lymphomas (PTCLs) encompass a heterogeneous group of clinically aggressive lymphomas with remarkable geographical variation in incidence1 (Figure 1A-B). PTCL incidence steadily increases with age and is higher in males than females, except for some PTCL subtypes that are more common in children, such as anaplastic lymphoma kinase (ALK)–positive anaplastic large cell lymphoma (ALK+ALCL), or in younger adults such as hepatosplenic T-cell lymphoma.2,3 The incidence of PTCLs has been increasing, amounting to 3.8% per annum in the United States,4 although it is not clear whether this is due to an actual increase or due to improved diagnosis. Although genetic and/or environmental factors are key factors in the pathogenesis of certain PTCLs such as human T-lymphotropic virus 1 HTLV-1) or Epstein-Barr Virus (EBV)–infected lymphomas and celiac disease associated intestinal lymphomas,1,5 most PTCLs are associated with genomic alterations occurring during normal T-cell development/differentiation or function.
Classification, incidence, and overall survival of PTCL subtypes. (A) Classification of peripheral T/NK cell lymphoma. PTCL classification is based on several parameters including clinical presentation, pathological, genetic features, and their association with normal cellular counterparts. Of the >30 PTCL entities recognized as distinct entities either by WHO or International Consensus Classification, the major subtypes present predominantly as nodal, extranodal, disseminated (leukemic), or cutaneous diseases, indicating that disease localization represents relevant diagnostic criteria for major PTCL entities. (B) Frequency of the PTCL entities. Epidemiological findings from published sources are reported (from North America [Bellei et al171 and Hsi et al172], the United States [Adams et al173 and Ruan et al174], South America [Fischer et al175], Africa [Fitzpatrick et al176 and Belarbi et al177], Western Europe [Laurent et al7], Central Europe [Janikova et al178], Nordic [Ellin et al8], India [Park et al179 and Nemani et al180], China [Park et al179 and Liu et al181], and Asia [Park et al179 and Yoon et al182]) (see citations in supplementary Table 2). The average frequencies from these studies were estimated and presented as pie charts for each specified region as indicated. ALK+ALCL vs ALK–ALCL cases were compared when data were reported. It must be noted that the frequencies presented may not represent the full spectrum of PTCL subtypes due to variations in data reporting across countries and also lack of requisite immunostains for current classification due to limited resources. Because of the exclusion of “other T-NHLs in individual regions or countries,” pie graphs may overrepresent the frequency of some PTCL subtypes. Different countries may have different reporting standards, subtype classifications, methodologies, or levels of data accuracy, which can affect the comprehensiveness and comparability of the data. However, for the worldwide frequency pie graph, the “other T-NHLs” were included to reflect the actual frequency of PTCL subtypes (see supplemental Table 2). (C) Overall survival of histological PTCL subtypes. These patients were generally treated with an anthracycline-containing regimen (adapted from Vose et al1). Regional 5-year survival is displayed in North America (Hsi et al172), Czech (Janikova et al178), and Asia (Yoon et al182). AITL, angioimmunoblastic T-cell lymphoma; CTCL, cutaneous T-cell lymphoma; EATL, enteropathy-associated T-cell lymphoma; HSTL, hepatosplenic T-cell lymphoma; MEITL, monomorphic epitheliotropic intestinal T-cell lymphoma; T-NHL, T-cell non-Hodgkin lymphoma.
Classification, incidence, and overall survival of PTCL subtypes. (A) Classification of peripheral T/NK cell lymphoma. PTCL classification is based on several parameters including clinical presentation, pathological, genetic features, and their association with normal cellular counterparts. Of the >30 PTCL entities recognized as distinct entities either by WHO or International Consensus Classification, the major subtypes present predominantly as nodal, extranodal, disseminated (leukemic), or cutaneous diseases, indicating that disease localization represents relevant diagnostic criteria for major PTCL entities. (B) Frequency of the PTCL entities. Epidemiological findings from published sources are reported (from North America [Bellei et al171 and Hsi et al172], the United States [Adams et al173 and Ruan et al174], South America [Fischer et al175], Africa [Fitzpatrick et al176 and Belarbi et al177], Western Europe [Laurent et al7], Central Europe [Janikova et al178], Nordic [Ellin et al8], India [Park et al179 and Nemani et al180], China [Park et al179 and Liu et al181], and Asia [Park et al179 and Yoon et al182]) (see citations in supplementary Table 2). The average frequencies from these studies were estimated and presented as pie charts for each specified region as indicated. ALK+ALCL vs ALK–ALCL cases were compared when data were reported. It must be noted that the frequencies presented may not represent the full spectrum of PTCL subtypes due to variations in data reporting across countries and also lack of requisite immunostains for current classification due to limited resources. Because of the exclusion of “other T-NHLs in individual regions or countries,” pie graphs may overrepresent the frequency of some PTCL subtypes. Different countries may have different reporting standards, subtype classifications, methodologies, or levels of data accuracy, which can affect the comprehensiveness and comparability of the data. However, for the worldwide frequency pie graph, the “other T-NHLs” were included to reflect the actual frequency of PTCL subtypes (see supplemental Table 2). (C) Overall survival of histological PTCL subtypes. These patients were generally treated with an anthracycline-containing regimen (adapted from Vose et al1). Regional 5-year survival is displayed in North America (Hsi et al172), Czech (Janikova et al178), and Asia (Yoon et al182). AITL, angioimmunoblastic T-cell lymphoma; CTCL, cutaneous T-cell lymphoma; EATL, enteropathy-associated T-cell lymphoma; HSTL, hepatosplenic T-cell lymphoma; MEITL, monomorphic epitheliotropic intestinal T-cell lymphoma; T-NHL, T-cell non-Hodgkin lymphoma.
Two classifications systems, the World Health Organization classification (WHO-HAEM5)6 and the new International Consensus Classification,2 use multiparametric approach for lymphoma diagnosis. PTCL classification is particularly challenging due to the rarity of the disease, wide morphologic spectrum, complexity of T-cell immunobiology, numerous T-cell functional subsets, functional modifications, and plasticity influenced by the microenvironmental niches.2,3,6 This is reflected in the discordance rate in PTCL diagnosis between referral and expert pathologists.7 Clinically, PTCLs are also hard to treat, with <30% surviving for 5 years.8 Although CHOP (cyclophosphamide, doxorubicin hydrochloride [hydroxydaunorubicin], Oncovin [vincristine sulfate], and prednisone)-based regimens remain the standard for first-line therapy, >75% of patients (excluding ALK+ALCL) fail to respond or relapse within 2 years (Figure 1C). Second-line agents such as histone deacetylase inhibitors and other agents have response rates <30% and poor progression-free survival9; thus, a better understanding of PTCL pathobiology is needed to improve diagnosis and treatment. In this review, we will highlight recent advances in the pathobiology of major PTCL subtypes and exciting directions that could be explored in the future.
Normal T-cell development, differentiation, and activation relevant to lymphomagenesis
The etiology of PTCL entities is affected by aberrations in critical genes/pathways regulating T-cell development, differentiation, and activation,10 and are concisely illustrated in Figures 2 and 3.
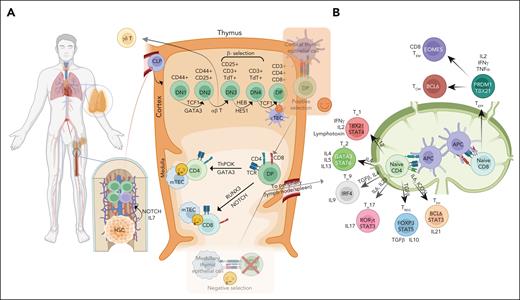
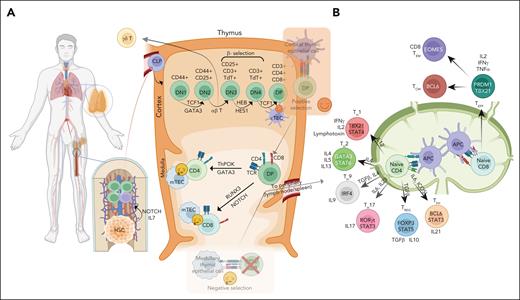
Stages of T-cell development and differentiation. (A) T-cell lymphopoiesis initiates with precursor HSC in the bone marrow (BM), the primary lymphoid organ; and CLPs in BM are directed to T-cell lineage fate via IL7/NOTCH signaling. CLP enters the cortex region of the thymus and undergoes a series of differentiation stages including β-selection and TCR repertoire phase, which can be identified through cell surface receptors (CD44 and CD25). T cells undergo a dual selection process, positive selection for compatibility of their TCR with self-MHC molecules and negative selection against autoantigenic peptides before leaving the thymus to form the peripheral T-cell repertoire. Double-positive cells differentiate toward CD4+ T cells through ThPOK and GATA3 expression, as well as a strong TCR signal, as well as toward CD8+ T cells through RUNX3 and NOTCH expression and a weak TCR signal. Although the vast majority of cells express an αβ TCR, a small subset expresses an alternative TCR composed of γ and δ chains (γδ T cells) and diverge early place at the DN3A stage in T-cell development and mostly become residents of mucosal sites. (B) In the peripheral secondary organs, the CD4+ T cells can differentiate into distinct TH subsets (TH1, TH2, TH9, TH17, Treg, and TFH), dependent upon stimulation from APC, their coreceptors, and cytokine milieu. Several PTCL subtypes show a similar cell state or transcriptomics associated with these TH subsets precursors. TH1 cells are characterized by the expression of key master regulators or TFs (ie, STAT4 and TBX21). These cells require interleukin-12 (IL-12) signaling for their differentiation and secrete IFN-γ, IL-2, and lymphotoxin. TH2 cells express TFs, STAT6 and GATA3, require IL-4, and produce IL-4, IL-5, and IL-13, important for TH2 differentiation. TH9 cells secrete IL-9, require TGF-β and IL-4, and are characterized by the expression of PU.1 and IRF4. TH17 expresses STAT3 and RORγt as key TFs, produces IL-17 and TNF-α, and requires IL-1β, TGF-β, IL-6, and IL-23. Tregs differentiate with TGF-β and IL-2, express FOXP3 and STAT5, and release TGF-β and IL-10 to suppress immune response. TFH cells produce IL21, with key TFs being BCL6 and STAT3, and require IL-6 signaling and ICOS-L. CD8+ T cells serve a cytotoxic function, regulated by PRMD1 and TBX21, and release IL-2, IFN-γ, and TNF-α. These cells can be further differentiated into CD8+ TEM and TCM via EOMES and BCL6, respectively. APC, antigen-presenting cell; CLP, common lymphoid progenitor cell; IFN-γ, interferon gamma; TCM, central memory T cell; TEM, effector memory T cell; TGF-β, transforming growth factor β; TNF-α; tumor necrosis factor α.
Stages of T-cell development and differentiation. (A) T-cell lymphopoiesis initiates with precursor HSC in the bone marrow (BM), the primary lymphoid organ; and CLPs in BM are directed to T-cell lineage fate via IL7/NOTCH signaling. CLP enters the cortex region of the thymus and undergoes a series of differentiation stages including β-selection and TCR repertoire phase, which can be identified through cell surface receptors (CD44 and CD25). T cells undergo a dual selection process, positive selection for compatibility of their TCR with self-MHC molecules and negative selection against autoantigenic peptides before leaving the thymus to form the peripheral T-cell repertoire. Double-positive cells differentiate toward CD4+ T cells through ThPOK and GATA3 expression, as well as a strong TCR signal, as well as toward CD8+ T cells through RUNX3 and NOTCH expression and a weak TCR signal. Although the vast majority of cells express an αβ TCR, a small subset expresses an alternative TCR composed of γ and δ chains (γδ T cells) and diverge early place at the DN3A stage in T-cell development and mostly become residents of mucosal sites. (B) In the peripheral secondary organs, the CD4+ T cells can differentiate into distinct TH subsets (TH1, TH2, TH9, TH17, Treg, and TFH), dependent upon stimulation from APC, their coreceptors, and cytokine milieu. Several PTCL subtypes show a similar cell state or transcriptomics associated with these TH subsets precursors. TH1 cells are characterized by the expression of key master regulators or TFs (ie, STAT4 and TBX21). These cells require interleukin-12 (IL-12) signaling for their differentiation and secrete IFN-γ, IL-2, and lymphotoxin. TH2 cells express TFs, STAT6 and GATA3, require IL-4, and produce IL-4, IL-5, and IL-13, important for TH2 differentiation. TH9 cells secrete IL-9, require TGF-β and IL-4, and are characterized by the expression of PU.1 and IRF4. TH17 expresses STAT3 and RORγt as key TFs, produces IL-17 and TNF-α, and requires IL-1β, TGF-β, IL-6, and IL-23. Tregs differentiate with TGF-β and IL-2, express FOXP3 and STAT5, and release TGF-β and IL-10 to suppress immune response. TFH cells produce IL21, with key TFs being BCL6 and STAT3, and require IL-6 signaling and ICOS-L. CD8+ T cells serve a cytotoxic function, regulated by PRMD1 and TBX21, and release IL-2, IFN-γ, and TNF-α. These cells can be further differentiated into CD8+ TEM and TCM via EOMES and BCL6, respectively. APC, antigen-presenting cell; CLP, common lymphoid progenitor cell; IFN-γ, interferon gamma; TCM, central memory T cell; TEM, effector memory T cell; TGF-β, transforming growth factor β; TNF-α; tumor necrosis factor α.
TCR signaling. T-cell activation is initiated by the interaction of TCR with peptide-MHC and coreceptors (ie, CD4 or CD8) and other costimulatory molecules including CD28, resulting in the multimolecular signalosomes, followed by the activation of several distal signaling pathways such as Ca2+-calcineurin-NFATs, PKCθ–IKK–NF-κβ, RASGRP1-RAS-ERK1/2, and TSC1/2-mTOR. The signaling complex containing scaffold adapter molecules, SLP-76 and GADs, which recruits PLCγ1 and tyrosine kinase ITKs. The tyrosine kinase ITK phosphorylates and activates PLCγ1, which then cleaves phosphatidylinositol (4,5) bisphosphate to generate 2 important second messengers, IP3 and DAG. IP3 binds to receptors on the ER, leading to an initial phase of calcium release, which subsequently activates the NFAT family of TFs, whose targets include many important cytokines, including IL-2 that are activated. Upon TCR activation, IL-2 receptor (IL-2R) activates JAK3, which in turn phosphorylates the beta-chain of IL-2R, recruiting JAK1. Both JAK1 and JAK3 in their active form phosphorylates (STAT), resulting in the nucleus transport. DAG activates several important proteins including several isoforms of PKC and RASGRP1 and RASGRP2, that are responsible for an initial phase of RAS activation, which is then sustained and amplified by SOS1. PKCθ binds to DAG and is recruited to the lipid rafts and triggers the formation of a trimolecular complex of adapter proteins in the cytoplasm called the CBM complex (CARMA1, BCL10, and MALT1), resulting in PKCθ–IKKβ–NF-κβ pathway activation. DAG induces the activation of another key molecule, RASGRP1, responsible for RAS activation in T cells, and initiates the RAS-MAPK cascade by activating the serine/threonine kinase Raf1 and activates MAPK ERK1 and 2. The costimulatory molecules CD28 mediates PI3K and VAV1 activation and further increases NF-κB and NFAT nuclear translocation, augmenting T-cell survival and production of the proliferative cytokine IL-2. Other TFs or pathways (p38MAPK, ERK1/2, and STAT3) are also crucial for T-cell activation and distinct functions associated with T cells. Several key genes involved in TCR are either genetically aberrant (highlighted in yellow text) or indirectly affected by other alterations in several PTCL subtypes as indicated below. Pathways that are relevant to PTCL entities are highlighted by brackets, followed by the PTCL entity in which the pathway is implicated in. CARMA1, caspase recruitment domain–containing membrane–associated guanylate kinase protein 1; DAG, diacylglycerol; ER, endoplasmic reticulum; ERK, extracellular signal–regulated kinase; IP3, inosine trisphosphate; MALT1, mucosa-associated lymphoid tissue translocation protein 1; mTOR, mammalian target of rapamycin; NFAT, nuclear factor of activated T cell; PKCθ, protein kinase C θ; PLCγ1, phospholipase Cγ1; RASGRP1, RAS guanyl nucleotide–releasing protein 1.
TCR signaling. T-cell activation is initiated by the interaction of TCR with peptide-MHC and coreceptors (ie, CD4 or CD8) and other costimulatory molecules including CD28, resulting in the multimolecular signalosomes, followed by the activation of several distal signaling pathways such as Ca2+-calcineurin-NFATs, PKCθ–IKK–NF-κβ, RASGRP1-RAS-ERK1/2, and TSC1/2-mTOR. The signaling complex containing scaffold adapter molecules, SLP-76 and GADs, which recruits PLCγ1 and tyrosine kinase ITKs. The tyrosine kinase ITK phosphorylates and activates PLCγ1, which then cleaves phosphatidylinositol (4,5) bisphosphate to generate 2 important second messengers, IP3 and DAG. IP3 binds to receptors on the ER, leading to an initial phase of calcium release, which subsequently activates the NFAT family of TFs, whose targets include many important cytokines, including IL-2 that are activated. Upon TCR activation, IL-2 receptor (IL-2R) activates JAK3, which in turn phosphorylates the beta-chain of IL-2R, recruiting JAK1. Both JAK1 and JAK3 in their active form phosphorylates (STAT), resulting in the nucleus transport. DAG activates several important proteins including several isoforms of PKC and RASGRP1 and RASGRP2, that are responsible for an initial phase of RAS activation, which is then sustained and amplified by SOS1. PKCθ binds to DAG and is recruited to the lipid rafts and triggers the formation of a trimolecular complex of adapter proteins in the cytoplasm called the CBM complex (CARMA1, BCL10, and MALT1), resulting in PKCθ–IKKβ–NF-κβ pathway activation. DAG induces the activation of another key molecule, RASGRP1, responsible for RAS activation in T cells, and initiates the RAS-MAPK cascade by activating the serine/threonine kinase Raf1 and activates MAPK ERK1 and 2. The costimulatory molecules CD28 mediates PI3K and VAV1 activation and further increases NF-κB and NFAT nuclear translocation, augmenting T-cell survival and production of the proliferative cytokine IL-2. Other TFs or pathways (p38MAPK, ERK1/2, and STAT3) are also crucial for T-cell activation and distinct functions associated with T cells. Several key genes involved in TCR are either genetically aberrant (highlighted in yellow text) or indirectly affected by other alterations in several PTCL subtypes as indicated below. Pathways that are relevant to PTCL entities are highlighted by brackets, followed by the PTCL entity in which the pathway is implicated in. CARMA1, caspase recruitment domain–containing membrane–associated guanylate kinase protein 1; DAG, diacylglycerol; ER, endoplasmic reticulum; ERK, extracellular signal–regulated kinase; IP3, inosine trisphosphate; MALT1, mucosa-associated lymphoid tissue translocation protein 1; mTOR, mammalian target of rapamycin; NFAT, nuclear factor of activated T cell; PKCθ, protein kinase C θ; PLCγ1, phospholipase Cγ1; RASGRP1, RAS guanyl nucleotide–releasing protein 1.
In brief, T-cell precursors derived from the hematopoietic stem cells (HSCs) migrate to the thymus for maturation and differentiation.11,12 The development of αβ T-cell precursors initiates with T-cell receptor β (TCR-β) locus rearrangement, followed by subsequent complex formation of TCR-β chain with pre–TCR-α,13 major histocompatibility complex (MHC) interaction, and the mature TCR-αβ receptor expression upon successful rearrangement.14 The helper (ie, CD4+) vs cytotoxic (CD8+) lineage choice is regulated by the antagonistic activities of master transcription factors (TFs), with ThPOK and GATA3 promoting CD4+ T-cell differentiation, whereas TFs such as RUNX3 promote CD8+ T-cell differentiation (additional details are available in previous studies15-17). γδ T cells mature in the thymus or extrathymic sites18 and represent 1% to 5% of T cells in the peripheral blood and 20% to 50% in mucosal sites,19 and this distribution also reflects the distribution of PTCL.
Upon cognate interaction with peptide-MHC complex, T cell undergoes clonal expansion, as well as activation and differentiate into effector and memory cells.20 The majority of the effector cells die when the initial stimulus is eliminated (ie, contraction phase), whereas the memory T cells persist for long periods.21 Although CD8+ T cells exhibit less functional diversity, CD4+ T cells further differentiate into regulatory T cells (Treg cells) or multiple helper T-cell (TH) subsets including TH1, TH2, TH9, TH17, and follicular helper T cells (TFH). The differentiation is governed by “master regulators,” such as B-cell lymphoma/leukemia 6 (BCL6) for TFH cells, TBX21 for TH1 cells, GATA3 for TH2 cells, and RORγδ for TH17 cells22 (Figure 2B).
T-cell activation is initiated with the cognate interaction of the TCR with peptide-MHC, facilitated by the interaction of the corresponding MHC with the CD4 or CD8 coreceptor and the engagement of the CD28 costimulator with CD80/CD86 on antigen-presenting cells, as illustrated in Figure 3. CD28 signaling and cytokines facilitate the signaling cascade, including phosphatidylinositol 3-kinase (PI3K) or JAK/STAT pathway for complete T-cell activation.23 Negative regulators attenuate the immune response either by competing with key coreceptors (eg, CTLA-4 against CD28)24,25 or by dampening the key signaling pathways such as PI3K-AKT via phosphatases (eg, PTEN, PHLPPL1/2, and SHIP1), negative regulators of the JAK/STAT pathway (eg, suppressor of cytokine signaling [SOCS] proteins), or guanosine triphosphatase (GTPase)-activating proteins (GAP) regulating GTPase activity and NF-κB pathway inhibitors.26 Immune checkpoint molecules such as programmed cell death protein 1 (PDCD1)/PD1, hepatitis A virus cellular receptor 2 (HAVCR2)/TIM3, T-cell immunoreceptor with Ig and ITIM domains (TIGIT), and lymphocyte activating 3b (LAG3) also limit T-cell reaction, and Fas cell surface death receptor/ligand interaction (eg, FAS/FASLG) induce cell death.26,27
In summary, normal T-cell differentiation and activation are well orchestrated by regulatory pathways. However, genetic and epigenetic aberrations may cause abnormal T-cell activation or regulation, an imbalance between survival and death, and failure of terminal differentiation, which can predispose to PTCL development. PTCLs can display characteristics of certain normal TH subsets, which could be derived from the corresponding committed memory TH cell (the cell of origin), but it is also plausible that the initial genetic alterations skew the polarization of the preneoplastic clone to a certain TH subset that becomes the feature of the eventual PTCL, thus reflecting the “differentiation state” of the PTCL (Figure 4).
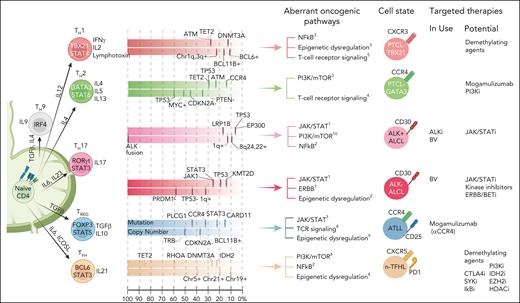
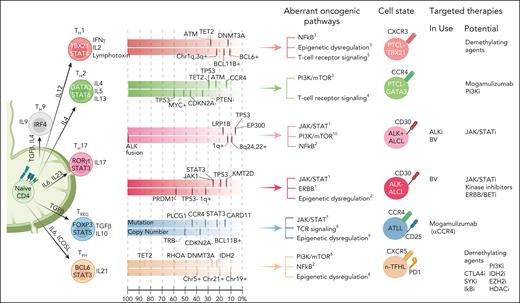
Major PTCL subtypes and association with TH cell states/cell-of-origin and genetic alteration. Although there is evidence from molecular or GEP studies indicating the association of transcriptional programs of major TH subset with PTCL subtypes, the genetic data suggest that specific driver genetic lesions may skew a naïve CD4+ T cell toward a TH differentiation to maintain that cell state and may take part with other genetic aberrations crucial for lymphomagenesis. It is also plausible that the genetic defects acquired along with transformation can modulate differentiation profiles and forced plasticity. Each colored box depicts mutation landscape and copy number data summarized from several recent genomic studies. The frequency of recurrent mutations/CN is arranged from left to right based on the frequency reported in the literature. Currently in-use and potential therapeutic targets are listed per cell state (Mereu et al,183 Feldman et al,184 Iqbal et al,38 Iqbal et al,10 Herek et al,158 Yu and Zhang,185 Sun et al,186 Liu et al,187 Bongiovanni et al,188 and Thakral et al189). ALKi, ALK inhibitor; BV, brentuximab vedotin; CN, copy number.
Major PTCL subtypes and association with TH cell states/cell-of-origin and genetic alteration. Although there is evidence from molecular or GEP studies indicating the association of transcriptional programs of major TH subset with PTCL subtypes, the genetic data suggest that specific driver genetic lesions may skew a naïve CD4+ T cell toward a TH differentiation to maintain that cell state and may take part with other genetic aberrations crucial for lymphomagenesis. It is also plausible that the genetic defects acquired along with transformation can modulate differentiation profiles and forced plasticity. Each colored box depicts mutation landscape and copy number data summarized from several recent genomic studies. The frequency of recurrent mutations/CN is arranged from left to right based on the frequency reported in the literature. Currently in-use and potential therapeutic targets are listed per cell state (Mereu et al,183 Feldman et al,184 Iqbal et al,38 Iqbal et al,10 Herek et al,158 Yu and Zhang,185 Sun et al,186 Liu et al,187 Bongiovanni et al,188 and Thakral et al189). ALKi, ALK inhibitor; BV, brentuximab vedotin; CN, copy number.
n-TFHL
The nodal lymphomas of TFH derivation (n-TFHLs) are considered a neoplasm of mature TFH cells with 3 subtypes under 1 umbrella, included in the WHO classification as angioimmunoblastic-type (AITL), n-TFHL, follicular type (n-TFHL-F), and n-TFHL, not otherwise specified (n-TFHL-NOS)2; or considered in the International Consensus Classification as AITL, T-follicular, and TFHL,NOS.6 The TFH immunophenotype is defined by the expression of 2 or more TFH markers (CD10, BCL6, CXCL13, PD1, and inducible T-cell costimulator [ICOS]).28 AITL is characterized by prominent vascular proliferation and follicular dendritic cell network3; n-TFHL-F is defined by a predominantly follicular growth pattern29,30; and n-TFHL-NOS has a different growth pattern that lacks the typical features of AITL. These subtypes share similar genetic landscapes, particularly mutations in epigenetic regulators,31 scattered to numerous B cells including large B-cell blasts, that are often infection by EBV.32 n-TFHLs may respond better to the histone deacetylase inhibitor or demethylating agent, such as 5-azacytidine.31,33,34
Regarding molecular pathogenesis we will focus mainly on AITL because most of the gene-expression profiling (GEP) studies include AITL, which demonstrated an association with a TFH cell signature, supporting the presumed cell of origin.35-37 n-TFHL-F and n-TFHL-NOS lack comprehensive genomic characterization, and defining the boundaries between n-TFHL-NOS and PTCL-NOS and assessing their biological differences require further large-scale systematic investigation.
The AITL GEP is dominated by a strong tumor microenvironment (TME) imprint, including overexpression of B-cell and follicular dendritic cell (FDC)-related genes, chemokines/chemokine receptors, and genes related to the extracellular matrix and vascular biology.10,36,38 The signature contributed by the neoplastic cells, albeit often quantitatively minor, is enriched in genes normally expressed by TFH cells, consistent with the cellular derivation of AITL (Figure 5). Several signaling pathways (eg, NF-κB, transforming growth factor β, and interleukin-6) are enriched in AITL compared with other PTCLs.35-37 An immunosuppressive TME, represented by high tumor-associated dendritic cell or macrophage gene signatures, is associated with inferior survival, whereas a high B-cell signature is associated with favorable clinical outcome.36
Gene expression–based molecular predictor for major PTCL entities. Amador et al107 used gene expression profiling for major PTCL subsets to differentiate each PTCL subtype including the novel molecular subtypes (PTCL-GATA3 and PTCL-TBX21). Shown are the expressions of known distinct transcripts for each patient in a molecular PTCL subtype (the detailed characteristics of these transcripts have been described in Amador et al107). Key histopathological features and immunohistochemistry biomarkers for each PTCL entity are shown under each molecular subtype.
Gene expression–based molecular predictor for major PTCL entities. Amador et al107 used gene expression profiling for major PTCL subsets to differentiate each PTCL subtype including the novel molecular subtypes (PTCL-GATA3 and PTCL-TBX21). Shown are the expressions of known distinct transcripts for each patient in a molecular PTCL subtype (the detailed characteristics of these transcripts have been described in Amador et al107). Key histopathological features and immunohistochemistry biomarkers for each PTCL entity are shown under each molecular subtype.
Genomic studies identified frequent aberrations dysregulating the epigenome (eg, TET2, DNMT3A, and IDH2R172) or proximal TCR/CD28 signaling pathway (eg, CD28 and PLCγ1) and RHOAG17V.39-45 The IDH2R172 mutations are distinctly associated with AITL and are rare in other n-TFHLs or PTCLs.39-41 These mutations produce the oncometabolite 2-R-hydoxyglutarate from α-ketoglutarate46 and inhibit α-ketoglutarate–dependent enzymes including the TET family. Although DNMT3A and/or IDH2 mutations co-occur with TET2 mutations in AITL, they are mutually exclusive in myeloid neoplasms.47,48 Interestingly, identical TET2 or DNMT3A mutations in neoplastic T cells may be detected in nonneoplastic cells (ie, myeloid and B cells) or HSCs, indicating their origin in HSCs. The identification of concurrent NOTCH1 mutations in the B cells may explain B-cell expansion through divergent clonal evolution in some AITLs.49,50 The clonal hematopoiesis associated with TET2 mutations may explain the increased incidence of myeloid neoplasm with TFH lymphoma.34,51 The second most frequent mutation, RHOAG17V, present in ∼50% AITLs,42,51-55 results in the glycine17→valine substitution and enhances interaction with vav guanine nucleotide exchange factor 1 (VAV1), leading to enhanced TCR signaling.56 This substitution may produce a dominant negative protein to decrease RHOA-guanosine triphosphatase activity, affecting ROCK function, and enhances RAC or PTEN activity, but this needs further investigation. Conditional murine models (eg, CD4+CreTet2–/–; RHOAG17V) cause abnormal Cd4+T-cell proliferation, aberrant TCR activation, and inactivation of FOXO1, inducing TFH differentiation and development of murine T-cell lymphoma.57-59 The TCR signaling pathway dysregulation due to CD28 mutations, a costimulatory receptor, and PLCγ1, a key intracellular effector, or other TCR pathway–related genes (VAV1, LCK, FYN, and ITK)39,42-45 is another major oncogenic mechanism in AITL. Fusion transcripts involving CD28 (ie, CTLA4::CD28 and ICOS::CD2839) enhance CD28 signaling60,61; other fusion proteins (ie, KHDRBS1::LCK and FYN::TRAF3IP2) enhance TCR–NF-κB activation,54,62 and the rare t(5;9)(q33;q22) translocation, resulting in interleukin-2 (IL-2)–inducible T-cell kinase (ITK)–spleen tyrosine kinase (SYK) (ITK::SYK) chimeric protein, induces a T-cell lymphoproliferation in mice,55,63,64 suggesting aberrant TCR signaling pathway activation in AITL pathogenesis. The IDH2R172 mutation also enhances TCR signaling,48 and the cooperative effect of Tet2 loss and IDH2R172 results in TFH-derived lymphoma in mice with dysregulated TCR signaling and germinal center B-cell interaction.65
ALCL
ALCL is a neoplasm composed of large/pleomorphic CD30+ anaplastic cells, involving lymph nodes often via sinusoidal pattern. Four distinct ALCL entities have been described, bearing distinct clinical and genetic features including ALK+ALCL, ALK−ALCL, primary-cutaneous ALCL, and breast implant–associated ALCL.68 The latter 2 entities have limited genomic studies and are summarized in supplemental Table 1, available on the Blood website.
ALK+ALCL represents 7% of PTCLs and is more frequent in North America than in Asia. ALK+ALCL shows male predominance,69 representing 20% of childhood lymphomas.70 The tumor cells often exhibit an aberrant T-cell immunophenotype (sometimes “null” immunophenotype) with defective expression of the TCR/CD3 complex and many pan T-cell antigens, despite a T-cell origin,71 which can be confirmed by clonal TCR gene rearrangements. They are strongly positive for CD30 on the cell membrane and Golgi area and have an aberrant ALK positivity, driven by ALK gene fusions, and often exhibit an activated cytotoxic immunophenotype, although the tumor tends to be CD4+. Although an anthracycline-based regimen remains the standard of care for ALK+ALCL, with a 5-year overall survival (OS) rate of 70% to 90%, younger age may be the predominant reason for better outcomes.72 Large retrospective series in adults reported that complete/unconfirmed complete response, partial response/stable disease, and progressive disease rates to the first-line therapy were 85%, 5%, and 10%, respectively, with etoposide showing beneficial effect, and the International Prognostic Index score is predictive of the progression-free survival and overall survival.69 The introduction of a chemoimmunotherapy regimen (ie, brentuximab-vedotin) has shown superior results in patients with CD30+ PTCL including ALCL.73
The majority of ALK+ALCLs show t(2;5)(p23;q35), resulting in ALK translocation (chromosome-2p23) with NPM1 (chromosome-5q35) and producing an NPM::ALK fusion protein with aberrant nuclear and cytoplasmic ALK expression and constitutive activation of the tyrosine kinase activity.74 However, many other partners lead to oncogenic ALK fusions.75 ALK represents the major oncogenic driver, and its pharmacologic inhibition has shown efficacy in relapsed/refractory patients with ALK+ALCL.76,77 The possible role of inflammation can be presumed from rare ALK+ALCL case reports presenting with skin lesions after an insect bite.78 The ALK-induced oncogenesis is mediated by engaging intracellular signals, including the JAK-STAT3,79 the RAS-extracellular signal–regulated kinase (ERK), the PI3K-AKT pathway,79 and other key effectors such as oncomiR-17∼92 cluster.80-82 In vitro studies suggest that increased PI3K signaling counteracted and PI3Kγ/δ inhibitor duvelisib potentiated crizotinib activity79,83 in ALK+ALCL cell lines.79,84 A transgenic murine model (Cd4;NPM-ALK), suggested an early thymic origin before TCR-β rearrangement and thymic β-selection is bypassed by NOTCH1 expression, although its relevance to ALCL is unclear.84
GEP analysis suggested a STAT3-regulated molecular profile,85 and messenger RNA (mRNA)–based diagnostic signatures included upregulated transcripts (ALK, TNFRSF8 [CD30], and TH17-cell–associated transcripts) and downregulated transcripts of TCR components36 (Figure 5). The TH17-like gene signature associated with ALK+ALCL is regulated by STAT386 and miR-135b.87 The AP-1 factors JUNB and BATF3 characteristically associate with ALCL transcriptional program,88 interact with interferon regulatory factor 4 (IRF4), and promote growth, survival, and ALCL phenotype.89 Moreover, BATFs bind classical AP-1 motifs, controlling T-cell growth and/or survival via MYC, CD30, and multiple lymphokines. Of note, the AP-1–BATF module promotes the expression of TH17, and JUNB and JUN modulate AKT1 transcription.90
ALK–ALCL encompasses ∼11% of PTCLs, with comparable morphology,1 albeit with larger and more pleomorphic cells than classical ALK+ALCL91 and with strong CD30 positivity and no ALK expression.1 ALK–ALCL occurs in older individuals, with less-frequent extranodal involvement and a prognosis inferior to ALK+ALCL.72
ALK–ALCLs show 6p25.3 rearrangements involving either IRF4 or DUSP22 with various partners (19%-30% cases).92 The TP63 rearrangement (7%-8%) due to inv(3)(q26q28) results in TP63-TBL1XR1 fusion and is mutually exclusive with DUSP22 rearrangement,92-94 although rare patients bearing both defects have been described.95 These genetic alterations are prognostic, with TP63 rearrangement having a worse prognosis, whereas DUSP22 rearrangement more often is associated with a good outcome,96-98 although some of these latter patients can display more aggressive courses.98-100 Cases with neither of the rearrangements have an intermediate prognosis.93,94 A subset of ALK–ALCL has oncogenic ERBB4-truncated transcripts carrying intronic 5' untranslated regions (UTRs)101 or recurrent mutation in MSCE116K or alteration in PRDM1/BLIMP1, 17p13(TP53), and CSMD2,102,103 with loss of TP53 and PRDM1 showing aggressive clinical course.94,104
ALCL can be distinguished from other PTCLs with a distinct set of upregulated transcripts (eg, TNFRSF8 [CD30], BATF3, and TMOD) and downregulated TCR-related transcripts.36,38,105,106 We described a 2-step algorithm to identify ALCLs from other PTCLs and the subsequent separation of ALK+ALCL from ALK–ALCL with high accuracy.106,107 GEP studies identified NF-κB and stromal signatures predictive of prognosis independent of ALK status in ALCL.108 ALK–ALCL shows higher genomic complexity than ALK+ALCL. Constitutive oncogenic JAK/STAT activation due to activating STAT3 and/or JAK1 mutations and recurrent gene fusions involving TFs (NFKB2 or NCOR2) and tyrosine kinases (ROS1 or TYK2) are frequent,103 suggesting that STAT3 activation represents a shared oncogenic mechanism in ALK+ALCL and subset of ALK–ALCL subtypes, exception include the DUSP22 rearranged subset.100,109
ATLL
Adult T-cell lymphoma/leukemia (ATLL) is a mature T-cell lymphoma caused by HTLV-1 and is endemic in southwest Japan, the Caribbean basin, Central and South America, and Central Africa. The average age at diagnosis is 40 to 60 years, but it is comparatively lower in Central and South America than in Japan.110 The disease is classified into 4 variants based on the degree of peripheral blood involvement, type of tissue infiltration, hypercalcemia, and lactate dehydrogenase levels.111
Foxp3 expression, a marker for Treg cells,112 led to the suggestion that ATLL cells may originate from HTLV-1–infected Treg cells. GEP studies demonstrated high expression of transcripts encoding IL2Rα, IL2Rβ, MHC-II, and proliferation-promoting pathways linked to HTLV-1 infection.113 We originally identified a unique mRNA signature, which included many of the transcripts induced by the TAX viral oncoprotein or involved in TCR signaling or associated with Treg cells (Figure 5).36 Interestingly, TAX expression in normal T cells is associated with increased H3K27me3, and there is a striking overlap between the induced peaks and the peaks found in ATLL.114 It is likely that many TAX-induced changes are epigenetically locked and persist even with the later repression of TAX expression. In contrast to TAX, the expression of the antisense RNA and corresponding HTLV-1 bZIP factor protein is quite universal, driving T-cell transformation and inducing proliferation, apoptotic resistance, multiple organ invasion, and drug resistance.115
Genetic studies showed that chromosomal break points tend to affect oncogenes and tumor suppressor genes and fragile sites with deletions of the NRXN3 locus (14q31.1 fragile site) in 60% of ATLLs.116 Activated TCR signaling observed in transcriptomic studies36,113 is likely due to mutations frequently affecting TCR/NF-kB pathways116 (Figure 3). CD28-mediated signaling is enhanced by several mechanisms, including focal gains and amplifications, mutations, and CD28 fusions with ICOS or CTLA4.117 Mutations in CCR4 (29%) and CCR7 (11%) usually result in the truncation of the intracellular C-terminal tail, prevent their internalization upon ligand stimulation, and enhance chemotaxis.118 Mogamulizumab, a monoclonal-CCR4 antibody, has shown efficacy in ATLL, particularly in cases with CCR4 mutation.119 Although. TP53 mutations are similar in Japanese and American patients, mutations of epigenetic and histone modifying genes (eg, EP300) are significantly higher in American ATLLs. In contrast, the mutation frequency in JAK/STAT and TCR/NF-κB pathway genes is significantly lower.120 ATLL also shows a CpG island DNA hypermethylator phenotype, resulting in inappropriate CpG islands (CGI) hypermethylation and inactivation of genes important in immune surveillance (eg, HLA-A, C). Interestingly, genetic alterations leading to 3' UTR disruption of CD274/PD-ligand 1 (PD-L1) result in decreased transcript degradation and increased protein expression,121 and alterations in B2M and CD58 are observed promoting immune evasion.
ENKTCL
Extranodal natural killer (NK)/T-cell lymphoma (ENKTCL) is an EBV-associated NK- or T-cell lymphoma, frequent in East Asia and Central and South America. It typically presents as destructive lesions in the nasal cavity, maxillary sinuses, or palate; and despite a localized presentation, it tends to relapse locally or at other extranodal sites. A minority of patients presents with extranasal disease, and they generally have worse prognosis compared with patients presenting with nasal disease (5-year survival, 50%). Aggressive NK-cell leukemia is also frequently EBV associated, with a similar geographical distribution as ENKTCL, but with limited molecular information122 and will not be further discussed in this section.
Most ENKTCL cases are derived from NK cells; however, ∼25% of the cases have TCR rearrangement and correspond to clonal neoplastic γδ or αβ T cells. The GEP signature of ENKTCL, irrespective of the cellular derivation, is distinctive and characterized by high expression of transcripts of NK-cell–associated receptors (eg, killer cell immunoglobulin- or lectin-like receptor [KIR or KLR]), cytotoxic markers (eg, GZMH and GZMB), distinct chemokine signatures, and TCRδ mRNA, but not CD16 (FCGR3A; Figure 5). The γδ-PTCLs from mucocutaneous sites are not EBV+ but showed a comparable gene expression profile with ENKTCL, suggesting that they are ontogenetically related and may share oncogenic pathways.36 Compared with normal NK cells, ENKTCL is characterized by activation of PDGFRA, JAK/STAT, MYC, AKT, and NF-κB pathways,123-125 and NK cell lines were sensitive to JAK/STAT3, aurora kinase, and NOTCH inhibition. NK cells can secrete many cytokines including interferon gamma, tumor necrosis factor α, granulocyte-macrophage colony-stimulating factor, and interleukin-10; and together with the upregulation of IP10/MIP2 chemokines in the TME, they may contribute to the occasional hemophagocytic syndrome; and together with cytotoxic effectors and FASL, they may mediate vascular damage and secondary necrosis.126
ENKTCL usually has a type II latency pattern of EBV infection, with the expression of EBERs, LMP-1, LMP2, and EBNA1. EBV is clonally present, but how EBV contributes to ENKTCL development is unclear, and the functional attributes of EBV components are derived mainly from B-cell investigations and may not be directly applicable to NK or T cells. Similar to some other EBV-associated malignancies, ENKTCL exhibits a CpG island DNA hypermethylation phenotype, leading to the inactivation of many tumor suppressor genes.127 Somatic mutations in genes regulating the JAK/STAT pathway, including STAT3 mutations, are the most frequent, oncogenic drivers.128 Mutations affecting DDX3X and TP53 are mutually exclusive, with each present in ∼10% to 15% of ENKTCLs. Although PRDM1 mutations are uncommon, 6q21 loss and promoter hypermethylation lead to frequent deficiencies in PRDM1 in >50% of cases.125,129 Gene mutations that may alter the epigenome are identified in 40% to 50% of patients and predominantly in chromatin modifiers such as histone lysine N-methyltransferases or demethylases (eg, KMT2D, KMT2C, and KDM2B), histone deacetylases (eg, BCOR, BCORL1, and NCOR1/2), histone acetyltransferases (eg, EP300 and CREBBP), the SWI/SNF (SWItch/sucrose nonfermentable) family members (eg, ARID1A and SMARCA4), and the DNA demethylase TET2. Importantly, genetic alterations in the 3' UTR of PD-L1, as described in ATLL, are also found in ENKTCL.121 Furthermore, frequent expression of PD-L1 transactivators, pSTAT3, interferon gamma, and HIF1α are noted in the tumor cells, which may contribute to the reported frequent PD-L1 expression.130,131 Interestingly, PD1/PD-L1 blockade has shown impressive efficacy in relapsed/refractory patients with ENKTCL.132 Two recent genetic studies128,133 attempted to define genetic subsets using mutation and copy number variations data, with certain subsets showing inferior prognosis.127,132 Xiong et al134 identified a minor subset enriched with T-cell signature and characterized by MGA mutation and BRDT loss, associated with a poorer outcome, whereas Dong et al121 identified a small cluster with poor prognosis, characterized by aberrations in genes affecting JAK3, BCOR, and TP53, and copy number abnormalities affecting chromosome 1 and 7. However, due to the retrospective nature of the above studies, lack of uniform treatment for patients with ENKTCL and less than vigorous NK- or T-cell lineage assignment, the biological significance of these novel genetic ENKTCL subtypes requires validation in further studies.
PTCL-NOS
PTCL-NOS is a heterogeneous group of predominantly nodal PTCL that do not correspond to any of the specific entities.135 Most cases are CD4+CD8–, but uncommon CD4–CD8+ and double-negative or double-positive cases are also observed. Most cases express the αβ TCR but uncommon γδ TCR or TCR-silent cases are seen. Importantly, neoplasms with a TFH immunophenotype are now excluded from PTCL-NOS, which removes ∼30% of the cases. Furthermore, most of the remaining cases can now be subdivided into lymphomas resembling different TH subsets or cytotoxic T cells, as detailed below. CD30 is often detected in a variable proportion of tumor cells. Up to 50% of PTCL-NOS contains an increased number of EBV+ B cells and is associated with poorer survival.134
The genetic features of PTCL-NOS are heterogeneous, and in studies without meaningful subgroup separation, results are difficult to interpret. Previously, cytogenetic analysis identified complex karyotypes, including gains of 7q and 8q,136 and 2 target genes on chromosome 7 (CDK6 on 7q22 and CARMA1 [caspase recruitment domain–containing membrane–associated guanylate kinase protein-1] on 7p22) have been implicated in PTCL-NOS pathogenesis.137,138 Chromosomal breaks involving the TCR gene loci (mostly the α/β-TCR locus at 14q11.2) are reported with translocation partner(s) identified in rare cases.136,137,139 The t(14;19)(q11;q13) involves the PVRL2 (poliovirus receptor–related 2) gene and induces overexpression of PVRL2 and BCL3 mRNAs.140,141 The IRF4 was identified as the partner of the TCRA locus in the t(6;14)(p25;q11.2) in 3 cytotoxic PTCL.142,143TP63 rearrangements are detected in <10% PTCL-NOS,144 and a recent genomic study identified TP53 and/or CDKN2A aberrant subgroup with marked genomic instability, associated with poor prognosis.145 Whole genome sequencing analysis identified distinct co-occurrence of CDKN2A and PTEN deletions in PTCL-NOS and demonstrated that the codeletion is distinctly associated with PTCL-NOS but not with other PTCL subtypes.146 Although there are limited studies on the PTCL-NOS mutational landscape, frequent mutations in epigenetic regulators,147FAT1148 and TP53,147 and genes associated with immune surveillance145 have been identified. VAV1 fusion transcripts are observe in 10% of PTCL-NOS, with several partners (VAV1::MYOF1, VAV1::THAP4, and VAV1::S100A7)149-151 having the common theme of eliminating the C-terminal SH3 autoregulatory domain and thus causing constitutive VAV1 activation.150 A small deletion affecting intron 25 extending to exon 26 of VAV1 is also described, resulting in an abnormal splicing variant, again leading to a constitutively active VAV1. Thus, VAV1 activation could be a significant pathogenetic mechanism in this group of lymphomas. Other rare but recurrent fusion transcripts involving FYN (FYN::TRAF3IP2), LCK (KHDRBS1::LCK),54,62 and CD28 (CD28::CTLA4,CD28::ICOS) 39 resulting in T-cell activation have been identified.152 Because TFH cases were not separated from PTCL-NOS in these studies, some of the mutations could be associated with the former. In future studies, it is essential to identify the different subtypes within the PTCL-NOS category so that the genetic landscapes can be clearly defined for each subtype and their association with prognosis can be reaffirmed (Figure 5).
Several initial GEP studies of PTCL-NOS identified distinct clusters of cases, but their biological significance was not clear and largely reflected the characteristics of the TME,153-156 activated CD4+ or CD8+ T-cell features,37 or CD8+ T-cell–like PTCL36 or cases enriched with NF-κB activation or proliferation signature.156,157 Using a large cohort of PTCL-NOS cases with rigorous pathology review and GEP analysis, we identified 2 novel molecular subgroups38 characterized by high expression of either TBX21 (TH1 master regulator) or GATA3 (TH2 master regulator) and corresponding target genes at both mRNA and protein levels (Figure 4). The “PTCL-GATA3” subgroup was associated with poor clinical outcomes and a lack of a prominent microenvironment signature. It was associated with high MYC and proliferation signatures; whereas the NF-kB pathway was enriched in the PTCL-TBX21 subgroup, which may explain the more favorable outcome associated with the NF-κB pathway reported previously.157 The PTCL-TBX21 subgroup (49% of PTCL-NOS) contains a subset with high cytotoxic signature, associated with poorer clinical outcome and enriched in mutations targeting DNMT3A methyltransferase domain.158 An independent study using GATA3 immunohistochemistry supported the poor clinical outcome of PTCL-GATA3 subgroup.159,160 Genomic copy number analysis revealed significant differences in genomic complexity between the 2 subgroups.161 PTCL-TBX21 has relatively lower genomic complexity and is associated with epigenetic dysregulation (TET2 and DNMT3A), whereas PTCL-GATA3 subgroup exhibited frequent del17p13 (TP53), del9p21.3 (CDKN2A), del6q21 (PRDM1), and del10p23 (PTEN).161 Notably, TP53 is often completely inactivated through del17p and/or mutation, but del10p23 (PTEN) is mostly heterozygous, and the codeletion of TP53 and PTEN is associated with poor prognosis.161 These findings are intriguing, because PTEN not only regulates PI3K-AKT activation162 but has partially overlapping functions with p53 for maintaining genomic integrity and stability.163-165 Due to their functional interdependency, the co-occurrence of these genetic aberrations is infrequent in hematologic malignancies or other cancers. Even in PTCL-GATA3, total loss of PTEN is rare, indicating that hyperactivation of PI3K/AKT pathway may be detrimental to mature T cells and/or PTEN serves some vital, non-PI3K–related functions that are difficult to compensate. Because targeting TP53 or CDKN2A aberrations are therapeutically challenging, their co-occurrence with the PTEN loss in PTCL-GATA3 subtype may be exploited for therapeutic vulnerability, because several clinical-grade inhibitors of PI3K pathways are available for clinical trials.166
Other rare extranodal PTCL entities are discussed in more detail by Iqbal and de Leval.167
Conclusion and relevance to precision medicine
Although current classification re-emphasizes the cell of origin or differentiation state of the T cell as an important criterion in classification and possible determinant of PTCL biology, high-throughput genomic and transcriptomic findings have revealed additional details in PTCL biology that may affect diagnosis and discover potential therapeutic targets of high clinical relevance168 (Figure 4). Epimutations, mutations affecting the proximal TCR signaling pathway, or more distal pathway activation extending to JAK/STAT signaling are characteristic of PTCL. T-cell activation is subject to the “Goldilocks” effect and needs to stay within certain limits to avoid detrimental effects, which applies to oncogenic TCR activation in the context of genetic and/or environmental perturbations. However, it is unclear under which in vivo conditions the preneoplastic T cells evolve, survive, and differentiate, requiring further in-depth investigation. If the cells are chronically stimulated, how they escape exhaustion and terminal differentiation are important areas of investigation. The TME is critically important for PTCL and has not been adequately studied. While mechanistic studies are ongoing, therapeutic interventions, especially targeting the epigenome or T-cell activation, show initial promise and should be further explored. We believe that accurate diagnosis and biological characterization of PTCL is a prerequisite for investigating precision treatments 169 or improving the efficacy with novel clinical trials.170 Of interest will be defining the biological boundaries between PTCL-NOS and other entities (eg, ALK–ALCL and n-TFHL-NOS) and further defining cytotoxic PTCL, so future clinical trials may be performed in the context of precise PTCL subgroups. As a rare disease, extensive international collaborative efforts are required to assemble sufficient cases for the above-mentioned biological studies and to discover vulnerabilities that may be targeted.
Acknowledgments
The authors acknowledge Dylan Jochum, Mehnaz Tabassum, and Alyssa Bouska in preparation of the manuscript. The authors thank many esteemed investigators in T-cell lymphoma biology whose work was cited in this article.
The work presented in this review from the authors’ laboratories was supported by the National Institutes of Health, National Cancer Institute (grants UH2/3 CA206127-02, R41CA221466-01A1, P01 CA229100 and R01CA251412 [J.I.]), and the Leukemia and Lymphoma Society (grant TRP-6129-04).
Authorship
Contribution: J.I. wrote the initial draft and planned the figures for the manuscript; W.C.C. and G.I. revised the sections relevant to the pathology and genetics of lymphoma subtypes (W.C.C. [n-TFHL, ATLL, ENKTCL, and PTCL-NOS] and G.I. [ALCL]); and J.I. and W.C.C. finalized the tables, figures, and the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Javeed Iqbal, Department of Pathology, Microbiology, and Immunology, James O. Armitage Center for Hematological Malignancies Research, University of Nebraska Medical Center, Omaha, NE 68198-3135; email: jiqbal@unmc.edu; Giorgio Inghirami, Pathology and Laboratory Medicine, Weill Cornell Medical College, 1300 York Ave, New York, NY 10065; email: ggi9001@med.cornell.edu; and Wing C. Chan, Department of Pathology, City of Hope National Medical Center, 1500 East Duarte Rd, Duarte, CA 91010; email: jochan@coh.org.
References
Author notes
The online version of this article contains a data supplement.


![Classification, incidence, and overall survival of PTCL subtypes. (A) Classification of peripheral T/NK cell lymphoma. PTCL classification is based on several parameters including clinical presentation, pathological, genetic features, and their association with normal cellular counterparts. Of the >30 PTCL entities recognized as distinct entities either by WHO or International Consensus Classification, the major subtypes present predominantly as nodal, extranodal, disseminated (leukemic), or cutaneous diseases, indicating that disease localization represents relevant diagnostic criteria for major PTCL entities. (B) Frequency of the PTCL entities. Epidemiological findings from published sources are reported (from North America [Bellei et al171 and Hsi et al172], the United States [Adams et al173 and Ruan et al174], South America [Fischer et al175], Africa [Fitzpatrick et al176 and Belarbi et al177], Western Europe [Laurent et al7], Central Europe [Janikova et al178], Nordic [Ellin et al8], India [Park et al179 and Nemani et al180], China [Park et al179 and Liu et al181], and Asia [Park et al179 and Yoon et al182]) (see citations in supplementary Table 2). The average frequencies from these studies were estimated and presented as pie charts for each specified region as indicated. ALK+ALCL vs ALK–ALCL cases were compared when data were reported. It must be noted that the frequencies presented may not represent the full spectrum of PTCL subtypes due to variations in data reporting across countries and also lack of requisite immunostains for current classification due to limited resources. Because of the exclusion of “other T-NHLs in individual regions or countries,” pie graphs may overrepresent the frequency of some PTCL subtypes. Different countries may have different reporting standards, subtype classifications, methodologies, or levels of data accuracy, which can affect the comprehensiveness and comparability of the data. However, for the worldwide frequency pie graph, the “other T-NHLs” were included to reflect the actual frequency of PTCL subtypes (see supplemental Table 2). (C) Overall survival of histological PTCL subtypes. These patients were generally treated with an anthracycline-containing regimen (adapted from Vose et al1). Regional 5-year survival is displayed in North America (Hsi et al172), Czech (Janikova et al178), and Asia (Yoon et al182). AITL, angioimmunoblastic T-cell lymphoma; CTCL, cutaneous T-cell lymphoma; EATL, enteropathy-associated T-cell lymphoma; HSTL, hepatosplenic T-cell lymphoma; MEITL, monomorphic epitheliotropic intestinal T-cell lymphoma; T-NHL, T-cell non-Hodgkin lymphoma.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/18/10.1182_blood.2023021787/2/m_blood_bld-2023-021787-c-gr1.jpeg?Expires=1767901530&Signature=Stb0DMrrHjc~DRyPQtG6yuafnvhDjJRrgKThoaWeXLE0vdMkDUgXjHxslVgRLOyX-t3Bcly09O5vEckacKBEBB3qsmchwEB5-daLgHeTARf3NmaH3M4e-MQJ4lmIkK8g0T6UK5S1VZka8cs6uDbfDEBBCC2MKm7NGC8Qf8XJ5nhFsSBU-v31ytFsEJPYWdZhHPbtNjd7Om8Wx59unMqyF-M5tWEXIaY9~~RXvyr6kWLandEv0LyGsCVGigKL9s6KbBiGJtXKivPKsuqg0cdMpBvquAXpgLbG48a7XqZ6GGEUTMfDjTmYTgncpIDJrD5TTG4GB8Uc5o7ATWyN2LIQGg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)




![Classification, incidence, and overall survival of PTCL subtypes. (A) Classification of peripheral T/NK cell lymphoma. PTCL classification is based on several parameters including clinical presentation, pathological, genetic features, and their association with normal cellular counterparts. Of the >30 PTCL entities recognized as distinct entities either by WHO or International Consensus Classification, the major subtypes present predominantly as nodal, extranodal, disseminated (leukemic), or cutaneous diseases, indicating that disease localization represents relevant diagnostic criteria for major PTCL entities. (B) Frequency of the PTCL entities. Epidemiological findings from published sources are reported (from North America [Bellei et al171 and Hsi et al172], the United States [Adams et al173 and Ruan et al174], South America [Fischer et al175], Africa [Fitzpatrick et al176 and Belarbi et al177], Western Europe [Laurent et al7], Central Europe [Janikova et al178], Nordic [Ellin et al8], India [Park et al179 and Nemani et al180], China [Park et al179 and Liu et al181], and Asia [Park et al179 and Yoon et al182]) (see citations in supplementary Table 2). The average frequencies from these studies were estimated and presented as pie charts for each specified region as indicated. ALK+ALCL vs ALK–ALCL cases were compared when data were reported. It must be noted that the frequencies presented may not represent the full spectrum of PTCL subtypes due to variations in data reporting across countries and also lack of requisite immunostains for current classification due to limited resources. Because of the exclusion of “other T-NHLs in individual regions or countries,” pie graphs may overrepresent the frequency of some PTCL subtypes. Different countries may have different reporting standards, subtype classifications, methodologies, or levels of data accuracy, which can affect the comprehensiveness and comparability of the data. However, for the worldwide frequency pie graph, the “other T-NHLs” were included to reflect the actual frequency of PTCL subtypes (see supplemental Table 2). (C) Overall survival of histological PTCL subtypes. These patients were generally treated with an anthracycline-containing regimen (adapted from Vose et al1). Regional 5-year survival is displayed in North America (Hsi et al172), Czech (Janikova et al178), and Asia (Yoon et al182). AITL, angioimmunoblastic T-cell lymphoma; CTCL, cutaneous T-cell lymphoma; EATL, enteropathy-associated T-cell lymphoma; HSTL, hepatosplenic T-cell lymphoma; MEITL, monomorphic epitheliotropic intestinal T-cell lymphoma; T-NHL, T-cell non-Hodgkin lymphoma.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/144/18/10.1182_blood.2023021787/2/m_blood_bld-2023-021787-c-gr1.jpeg?Expires=1767901531&Signature=ZEFQx~RxhHkIKFk2go2EERfnO5Dsq1PbN9FkxVzOP9JXvRyNey89ruHq-2V5N-zWITJz6xz8yv8NyMvDxjqa46U6wPgLq79ap~dLE6nLHOxn-XOgrJP9Yx3D9XUQY3gFND1zlrjxAXg5mS6haRlUsUUzO02akUF4z~5RgY2wu3CGYA188R-cJwifKkRJmb-g4E-TKbFmCgJU~gsSkF5ygdLarWmSBk6DclhjUl91rTw9jlJbRjsGnhydO9WfgmjiNx8SyvvsGPIDinjiEF9QOmWBmK5Fh6Bt~bCYMDD7mE0DVp2bpkbB1FFRZSSwiqGHIVuJSVx~nFHFpqP0JJlZrQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)