In this issue of Blood, Sureda-Gómez and colleagues provided pivotal insights into the molecular subtyping of Burkitt lymphoma (BL).1 In particular, they demonstrated, for the first time, the mutual exclusivity of SRY-related HMG-box gene 11 (SOX11) expression and Epstein-Barr virus (EBV) presence in BL cases. This novel observation could serve per se as a new biomarker for subclassifying BL and supports the most recent classification of BL, based on EBV status.2 The exploration of the SOX11 in the context of BL is particularly intriguing given its established oncogenic role in mantle cell lymphoma (MCL) and absence in normal B cells and other B-cell lymphomas.3
In the first part of their study, the authors reviewed a large series of BL cases for which molecular and clinical data were available. Based on RNA-sequencing data, as well as immunohistochemistry, they observed that only a subset of EBV− cases, but no EBV+ BL case, expressed SOX11. Accordingly, they divided BL into 3 categories, namely EBV−SOX11+, EBV−SOX11− (“double negative”), and EBV+SOX11−. Of note, among the EBV− BL, SOX11 expression status was associated with significant genetic differences, suggesting a possible distinct molecular pathogenesis.
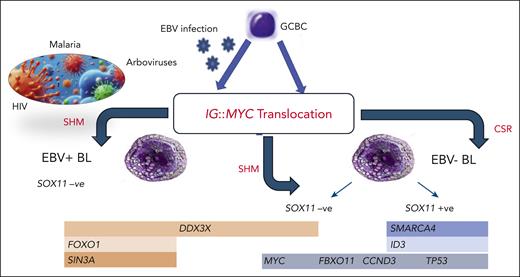
In fact, first of all, by analyzing the breakpoint regions of the IG∷MYC translocations, the authors found that the genetic aberration is predominantly generated by class switch recombination in EBV−/SOX11+ BL and by somatic hypermutation in EBV−/SOX11− BL. This suggests that SOX11 status is associated with the early pathogenetic event of MYC translocation.
In addition, a different somatic mutation pattern was observed in the 3 subgroups. Specifically, a higher incidence of SMARCA4 and ID3 mutations was recorded in SOX11+ BL. This indicated that not only the presence of EBV but also the SOX11 status is associated with a peculiar mutational pattern in BL (see figure).4
FBL classification based on the molecular pathogenesis. The major driving event, the IG::MYC translocation, may occur in either EBV+ or EBV− germinal center B-cell (GCBC, presumably an early centroblast). Thereafter, different additional genetic lesions (overall more abundant in EBV− cases) would contribute to BL phenotype. In EBV− cases, a major role seems to be played by SOX11 regulation, which is associated with specific gene expression as well as a mutational profile. Apparently, also the IG::MYC translocation itself is mediated by different mechanisms, namely somatic hypermutation (SHM) and class switch recombination (CSR), according to the EBV and SOX11 status.
FBL classification based on the molecular pathogenesis. The major driving event, the IG::MYC translocation, may occur in either EBV+ or EBV− germinal center B-cell (GCBC, presumably an early centroblast). Thereafter, different additional genetic lesions (overall more abundant in EBV− cases) would contribute to BL phenotype. In EBV− cases, a major role seems to be played by SOX11 regulation, which is associated with specific gene expression as well as a mutational profile. Apparently, also the IG::MYC translocation itself is mediated by different mechanisms, namely somatic hypermutation (SHM) and class switch recombination (CSR), according to the EBV and SOX11 status.
To get insights into the possible role of SOX11 in BL, functional experiments were carried out on different BL cell lines, induced to express SOX11 or knocked out by CRISPR-Cas9 to suppress its expression. When gene expression profiles were generated and studied to compare SOX11+ and SOX11− samples, a molecular signature of 79 genes was identified. Intriguingly, the authors also investigated whether SOX11 may act similarly in BL and MCL. They found that in both diseases SOX11 may contribute to deregulation of oxidative stress, heterotrimeric G proteins, chemokines and cytokines, integrins, angiogenesis, and PDGF signaling pathways. It is worth noting that despite the partial overlap with the transcriptional program of MCL, SOX11+ BL cells did not show differences in cell migration or B-cell receptor (BCR) signaling compared with their SOX11− counterparts. This finding suggests that SOX11 may play, at least in part, a different role in BL than in MCL, acting through other pathways or cellular processes. The increased adhesion to VCAM-1 observed in SOX11+ BL provides a glimpse into these alternative mechanisms, which could affect tumor microenvironment interactions and possibly tumor dissemination or localization.
In this regard, the study’s emphasis on the SOX11-associated gene expression profile in BL also underscores the power of next-generation sequencing and RNA-sequencing technologies to uncover the complexities of cancer. As we delve deeper into the molecular characterization of lymphomas, we can begin to tailor treatments more effectively. For instance, patients with BL exhibiting SOX11 overexpression might benefit from novel therapies targeting the specific pathways altered in this subset of BL, potentially improving outcomes. Despite the remarkable cure rate that can exceed 80%, relapsed or refractory BL cases are still characterized by an extremely poor prognosis, and no treatment or agent has been approved specifically for this indication. Targeting the BCR signaling as well as the use of the newest bispecific antibodies and CAR-T5 should be considered, but the rarity of the disease limits our capability to design clinical trials.
In this study, it was also shown that the number of BL, and more generally of lymphomas (and maybe cancers) that are associated with EBV infection, is likely underestimated. The ultrasensitive investigation of EBV in 37 BL samples that were negative for EBER did reveal that 12 of 37 were positive for BamHI-W, of which 9 were positive for EBNA1. Of the 12 cases, 10 were SOX11−. This is in line with previous observations made by microRNA analysis and supported once more the so-called hit-and-run hypothesis.6
However, despite the fact that the recent classification based on EBV status is fully representative of BL molecular profiles, as largely indicated by gene expression as well as mutational analyses,4,7-10 the full understanding of BL pathogenesis should not forget the epidemiological features that supported the historical subclassification of the disease (endemic, sporadic, and immunodeficiency associated). For example, EBV and stochastic mutations do not fully explain the high frequency of the disease in the periequatorial belt with the association with malaria and arboviruses infection. HIV infection, the second commonest setting for BL development, cannot be reduced to EBV emergence only, as chronic antigen stimulation and possible contribution of other pathogens (including HIV itself) cannot be ruled out.
In conclusion, this article contributes significantly to the field of lymphoma research by unveiling the distinct molecular subsets within EBV− BL, based on SOX11 expression. It provides further evidence for the subdivision of BL based on molecular characteristics, thus refining our approach to diagnosis and treatment. The mutual exclusion of SOX11 and EBV, along with the distinct genetic features associated with SOX11 expression, suggest that even within relatively homogenous disease (like BL), molecular subtypes do exist. This could have far-reaching consequences for patient stratification, prognostic assessment, and the development of individualized therapies that exploit the unique vulnerabilities of each BL subset.
Conflict-of-interest disclosure: The author declares no competing financial interests.